Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations—a review
Introduction
Lung cancer is the leading cause of cancer related deaths in Canada (1). In the developed world, non-small cell lung cancer (NSCLC) is the predominant form of the disease, accounting for approximately 85% of cases (2). The advent of molecular profiling has led to the discovery of “driver mutations”, targeted therapy, and personalized medicine. Some of the earliest driver mutations discovered and targeted were mutations in the epidermal growth factor receptor (EGFR) gene (Figure 1). EGFR is a receptor tyrosine kinase which, once activated by binding ligand and receptor dimerization, transphosphorylates its cytoplasmic tails, activating cellular signaling pathways such as the phosphoinositide 3-kinase (PI3K)-AKT pathway, the STAT pathway, and the MAPK pathway, ultimately leading to increased cell proliferation, migration, and survival (3-6). Approximately 10-30% of NSCLC patients have activating mutations in EGFR (7-9). Targeting EGFR in these patients with activating mutations has shown initial and significant success in the clinic (10,11).
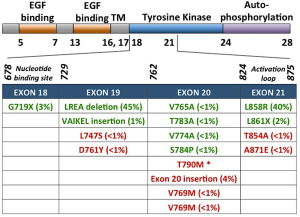
Classical activating mutations, such as the exon 19 deletions and exon 21 L858R substitution, account for approximately 45% and 40% of all EGFR mutations, respectively; these two mutations are associated with good responses to EGFR-targeted small molecule inhibitor therapies (11). Initially, these mutations were shown to destabilize the auto-inhibited conformation of the receptor (the normal state of the receptor in the absence of ligand) thus causing constitutive activation of the kinase domain (12-14). More recently, Shan et al. (15) reported that the L858R mutation causes a partially disordered state of the EGFR kinase which promotes dimerization and thus aberrant activation. Dixit and Verkhivker (16) recently published the sequence and structure-based computational model which predicted that the L858R mutation synergistically shifts EGFR towards the active state and favours the formation of the asymmetric dimer. The L858R activating mutation has also been shown to decrease ATP binding affinity. Yun et al. (17) report that this decreased affinity for ATP essentially creates a “therapeutic window”, which renders the oncogenic EGFR mutants more easily inhibited by TKIs, as they now have higher binding affinity than, and thus can outcompete ATP.
Over the years, drugs have been developed which specifically target EGFR. One such class is a group of small molecule inhibitors that inhibit the tyrosine kinase domain of EGFR, and are thus referred to as tyrosine kinase inhibitors (TKIs). The first TKIs shown to have clinical benefit were gefitinib and erlotinib (10,11,18). These two TKIs are considered first-generation; they reversibly bind to the tyrosine kinase domain of EGFR (19). First-generation EGFR TKIs have shown significant success clinically in patients with the most common activating EGFR mutations. As first-line treatments, EGFR inhibitors have been shown to produce overall response rates (ORRs) of close to 75% in patients who harbor activating mutations in EGFR (3,20,21).
Despite this, the vast majority of patients develop resistance to treatment; the median progression free survival (PFS) after treatment with a first generation EGFR TKI in patients with activating mutations is typically less than one year (20-22). Numerous biological mechanisms of acquired resistance (AR) have been elucidated (Figure 2), but in up to 30% of patients, the mechanism of resistance remains unknown (23). To date few patients have been cured by an EGFR TKI alone and almost all patients eventually acquire resistance and relapse (21,24). This review aims to give an overview of the most common mechanisms of primary and AR as well as highlight novel, newly emerging theories.
Primary resistance
EGFR somatic mutations
Depending on the mutation present in EGFR, tumors exhibit differential TKI sensitivities. While the most common EGFR-activating mutations, L858R and exon 19 deletion, typically confer sensitivity to EGFR TKIs, other primary EGFR mutations can confer resistance. Exon 20 insertions or duplications, which account for approximately 4-9% of EGFR mutations, appear to be resistant to EGFR inhibitors in vivo, despite the fact that these mutations appear to also be activating mutations, at least in vitro (25-33). Most of these insertions occur between amino acids 767 to 774 (31). The crystal structure of the exon 20 D770_N771insNPG EGFR mutant revealed that the ATP-binding pocket is unaltered, thus EGFR is activated without increasing its affinity for EGFR TKIs (34). Interestingly, loss of these activating EGFR mutant genes has been reported in vitro, which leads to a decrease in addition to EGFR signaling, gained addiction to both HER2/HER3 and PI3K/AKT signaling, and thus AR to EGFR TKI (35). Other, much less frequent, primary EGFR mutations such as G719X and L861X, have been reported (Figure 1) (36,37).
Although recognized mainly as a mechanism for AR, another EGFR exon 20 mutation, T790M, has also been associated with primary resistance. This mutation is within the gatekeeper residue, and restores the L858R mutant receptors affinity for ATP to wild-type levels, thus decreasing the effect of TKIs (38). Biochemical studies have demonstrated synergistic kinase activity and transformational potential when T790M is concurrently expressed with a TKI-sensitizing, EGFR-activating mutation (39,40).
Minor clones with the T790M mutation have been identified in treatment-naive tumors that contain classic sensitizing mutations. While this mutation has low allelic frequencies in treatment-naive tumors, pressure from TKIs may select for enriched growth of these T790M clones, leading to overall AR. As allelic dilution most likely obscures the detection of de novo T790M mutations via conventional Sanger sequencing methods, higher sensitivity assays such as high-performance liquid chromatography, mass spectrometry, locked nucleic acid PCR techniques and next generation sequencing have been suggested as alternate screening methods (41-47). Recent studies using these more sensitive techniques have reported T790M mutations in 35%, 38%, and 79% of EGFR-mutant, NSCLC pretreatment specimens (48-50). Interestingly, Rosell et al. (48) reported that low levels of BRCA-1 negates the desensitizing effects of the T790M mutations and is associated with longer PFS to erlotinib. Conversely, high levels of BRCA-1 lead to increased DNA damage repair capacity and thus de novo resistance.
EGFR germ line polymorphisms associated with primary resistance
T790M
This mutation has also been identified rarely in patients as a germline polymorphism; it has been identified in 0.5% of never smoker-lung cancer patients’ blood samples (51). Furthermore, the T790M mutation has also been putatively associated with familial cancer syndromes (52). In short, the proband’s mother, maternal grandfather and great uncle all succumbed to bronchioloalveolar carcinoma in their 60’s and 70’s. Furthermore, three out of the four siblings, including the proband, also developed lung cancer; two of these individuals (including the proband) failed to respond to gefitinib treatment, alone or in combination with chemotherapy. The third sibling was only recently diagnosed at the time of the referenced publication, thus their cancer treatment and subsequent response were not reported. Tumor specimens were available from two of the siblings (five independent primary tumors from the proband and a biopsy from metastatic disease from a brother). EGFR sequencing identified the T790M mutation in all tumors in a 1:1 ratio with the wild-type allele. Three of the five tumors from the proband had additional EGFR somatic mutations that typically respond to EGFR TKI therapy (two with L858R, one with delL747_T751); the biopsies from the remaining two primary tumors revealed no additional mutations in EGFR. The biopsy from the brother’s metastatic lesion also harbored the G719A EGFR mutation, which typically confers sensitivity to EGFR TKI therapy. Most intriguingly, the T790M mutation was also present in the germline (measured from peripheral blood mononuclear cells) of both individuals as well as their other two siblings (52). In the report by Girard et al. (51), no response information to EGFR TKI was reported.
V843I
In 2008, there was a care report about a woman with a family history of lung cancer (father and a brother) who was diagnosed with multiple adenocarcinomas that exhibited either L858R or L861Q EGFR mutations as well as a rare germline EGFR mutation, V843I. Three of her four remaining siblings were sequenced, two of whom also harbored the germline mutation, neither of whom had developed lung cancer despite their advanced age (67 and 72 years of age) (53). Another report was published in 2011 on a family with a history of cancer where four of the family members exhibited the germline V843I mutation (54). Three of these family members developed lung cancer, and all of them had the EGFR somatic L858R mutation. Only the proband underwent EGFR TKI therapy, however they did not respond to either gefitinib or erlotinib. The most recent report of this germline variation was in 2013, which described the first Caucasian patient with this mutation as well as the first patient without concomitant additional known EGFR-activating mutation (55). This patient did not respond to erlotinib and their tumors continued to grow rapidly while on this treatment. Modeling analysis of V843I suggests that ATP and TKI affinities for EGFR are not affected by this mutation; the mechanism of action for a possible germ line predisposition of V843I to develop lung cancer remains unknown. Matsushima et al. (56) demonstrated that the V843I mutation increased the phosphorylation of EGFR and downstream signaling proteins compared to wild type EGFR, especially when induced by EGF, suggesting a potentially oncogenic role for this mutation. Furthermore, they demonstrated that the double V843I/L858R mutant did not have increased phosphorylation levels, however the double mutant was resistant to erlotinib, gefitinib, afatinib and dacomitinib. Finally, structural modeling suggests that TKI binding to EGFR would be sterically hindered by Arg841 in the V843I/L858R double mutant (56).
Other genetic polymorphisms
BIM
Despite our furthered understanding of the sensitizing effects that various EGFR mutations have to TKIs, patients with identical mutations can demonstrate a spectrum of responses. One explanation for this variability in responses lies within the apoptotic machinery. Recent studies have demonstrated up-regulation of BIM in response to EGFR TKIs in mutant cell lines, which correlated with apoptotic response. EGFR-mutant patients with low BIM expression prior to treatment exhibited less tumor shrinkage and shorter PFS after TKI therapy (57-61). Variances in BIM expression levels have been suggested to be due to a genetic polymorphism in BIM, leading to alternative splicing and altered function (58,59,62,63). Clinically, the BIM deletion polymorphism has been reported in 12.9% of East Asian individuals. Furthermore, patients with NSCLC who harbor this BIM polymorphism exhibit significantly inferior responses to EGFR-TKI treatments compared to wild-type BIM counterparts (64). Indeed, Nakagawa et al. (64) demonstrated sensitization in EGFR-TKI resistant cell lines that harbor BIM polymorphisms by combination therapy with HDAC inhibitor vorinostat. Recent results from the randomized phase III EURTAC trial demonstrated that high BIM expression prior to treatment was a marker of longer PFS (HR =0.49; P=0.0122) and overall survival (HR =0.53; P=0.0323) (65). As such, BIM appears to act as both a biomarker and mediator of TKI-induced sensitivities in several oncogene-driven cancers.
Acquired resistance (AR)
Secondary EGFR mutations
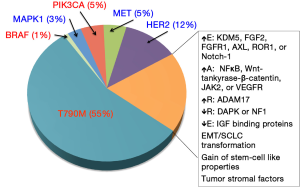
The earliest reported mechanism of resistance to TKIs in EGFR-mutant NSCLC is the T790M mutation (see previous section on primary resistance), which accounts for approximately 50-60% of cases with AR to EGFR TKI therapy (24,66-69). Despite the multiple avenues of enhanced oncogenicity, tumors harboring T790M mutations often exhibit surprisingly slow growth rates (70). A retrospective study examining T790M status on rebiopsy specimens from 93 patients with EGFR-mutant lung cancer and AR to TKIs found that T790M patients had a better prognosis. Furthermore, lack of T790M at time of rebiopsy was associated with a poorer performance status at progression, earlier development of new metastatic disease sites, as well as shorter post-progression survival (24).
Other secondary mutations in EGFR linked to AR have also been identified such as D761Y, T854A, and L747S. However, the structural basis for how these mutations confer resistance remains unknown (71-73).
Gene copy alterations of alternative pathways
MET
Amplification of the MET gene is considered one of the more common causes of AR in EGFR-mutant NSCLC. Heterodimerization of MET and ERBB3 leads to sustained activation of the PI3K/AKT signaling pathway, bypassing the inhibition of EGFR conferred by TKIs (74). Initial reports suggested that MET amplification accounted for approximately 22% of AR cases, independent of T790M status. However, two recent studies, each testing 37 patients with AR to EGFR TKIs for MET amplification by FISH, suggest that this prevalence is closer to 5% (44,75). This discrepancy between studies may be in part due to technical difficulties in identifying this genetic alteration in clinical samples. The initial studies with the higher reported percentage of MET amplification used several methods of assessment such as array comparative genomic hybridization (aCGH), quantitative real-time PCR, as well as FISH. On its own, FISH is the most widely acceptable technique in clinical laboratories, however technical difficulties arise due to both MET and EGFR being on chromosome 7. Furthermore, polysomy of chromosome 7 is common in NSCLC, particularly in samples with EGFR activating mutations (76). As such, it’s been suggested that new clinical protocols to distinguish meaningful MET amplification and copy number gain from underlying polysomy in both EGFR-mutant and wild-type lung cancers, is required. Aberrant activation of MET and subsequent AR has also been reported via excessive hepatocyte growth factor secretion, the natural ligand for MET (77,78). MET-amplification may not be solely a mechanism of AR but also an inherent event. Low frequencies of MET-amplified subclones have been identified in treatment naive specimens (79). Similar to the development of AR in tumors with low frequencies of T790M, the dominant mechanisms of AR at the time of disease progression in the majority of these cases has been MET amplification (80). Recent and on-going attempts to overcome AR due to overriding EGFR inhibition via aberrant MET signaling is to inhibit both receptors simultaneously (80-83). Overall, there is reasonable rationale for clinical trials to evaluate MET inhibitors in patients who developed AR to EGFR TKI therapy via MET amplification mechanism.
HER2 amplification
Recently, amplification of HER2 has been reported in three of 26 (12%) EGFR-mutant NSCLC patients who have AR to TKIs. Similar to MET, it is believed that HER2 is able to signal parallel to inhibited EGFR and thus reactivate common downstream signaling pathways (84).
MAPK amplification
Due to KRAS mutations’ associations with primary resistance to EGFR inhibitors, recent studies have focused on RAS/MAPK signaling as potential mechanisms of AR (85). KRAS mutations themselves are known to be mutually exclusive with EGFR mutations in patients. Thus, despite their role in primary resistance, no KRAS mutations have been identified in EGFR mutant patients with AR (75,85,86). However, Ercan et al. (87) identified MAPK1 amplification in an erlotinib-resistant EGFR-mutant NSCLC patient. The investigators further demonstrated that a mechanism of resistance to the irreversible EGFR TKI WZ4002 was increased ERK signaling due to amplification of MAPK or down regulation of negative regulators of ERK signaling. This resistance was overcome by inhibition of MEK or ERK and prevented the development of subsequent resistance.
Mutations in downstream effector molecules of EGFR
PIK3CA mutations
Alternative to parallel pathways being activated, downstream effector molecules of the EGFR signaling pathway have also been reported to be mutated, leading to AR (76). PIK3CA mutations have been reported in 5% of EGFR-mutant patients who have AR and preclinical studies demonstrate the ability of these mutations to confer resistance via activation of downstream AKT (88). PI3K phosphorylates PIP2 to PIP3; PTEN (phosphatase and tensin homolog), reverses this phosphorylation. The loss or decreased expression of PTEN has also been linked to AR (89,90).
BRAF mutations
A recent retrospective study identified point mutations in BRAF in two out of 195 (1%) lung cancer patients with AR to EGFR TKIs. The investigators further confirmed BRAFs potential role in AR by inducing ectopic expression of mutant BRAF in drug-sensitive EGFR-mutant cells, inducing resistance to EGFR TKIs. The addition of a MEK inhibitor was able to overcome induced resistance (86).
Epigenetic and other mechanisms
Epigenetic
Although the genetic basis for acquiring TKI resistance has been well established, a number of recent observations reveal a reversible epigenetic mechanism of drug resistance. Firstly, genetic mechanisms alone cannot account for the high prevalence of TKI-resistant tumors. Secondly, many NSCLC patients who previously developed TKI resistance respond to TKI again after being off the drug for a period of time. Such a phenomenon indicates that acquired TKI resistance might not require a permanent genetic alteration. Thirdly, there is still a significant proportion of TKI resistant tumors that do not harbor any known genetic alterations and activation of alternative signaling pathway. Finally, tumors exhibit not only genetic but also epigenetic heterogeneity within cell populations (91,92).
Epithelial-to-mesenchymal transition (EMT)
EMT, as the name suggests, is a cellular phenotypic change. It can be characterized molecularly by a loss of epithelial markers such as E-cadherin, and a gain of mesenchymal markers, such as vimentin (93). At the cellular level, EMT leads to enhanced motility, invasiveness, and in vitro EGFR TKI resistance (94-96). EMT has also been identified in subsets of clinical EGFR TKI-resistant specimens. Despite the growing evidence that EMT may play a role in resistance to treatments, the underlying biology of this change and specific mechanisms of resistance remain unknown (75). Recent work demonstrated the efficacy of blocking ERK1/2 in preventing EMT in lung cancer cells and enhancing their sensitivity to EGFR TKIs. By inhibiting MEK1/2 (MAPKK1/2), an epithelial phenotype was promoted and maintained in NSCLC cells despite exogenous stimulation by TGF-beta. Furthermore, cells that exhibited de novo or AR to gefitinib demonstrated decreased cell migration and enhanced sensitivity to the EGFR TKI when MEK was inhibited long enough to trigger changes in EMT marker expression (97).
Histological transformation
Several studies have reported the histological transformation to small cell lung cancer in EGFR mutant NSCLC patients with acquired EGFR TKI resistance, accounting for resistance in possibly up to 3% of the patients. Interestingly, the conversion to SCLC was associated with sensitivity to standard SCLC treatment while the original EGFR mutation was still maintained in the tumor (75,98). The mechanism underlying this histological transformation still remains unknown.
AXL activation
AXL is a tyrosine kinase receptor which induces cell proliferation, migration and invasion in cancer. Recently, several groups reported that activation of AXL signaling pathway may confer TKI resistance in EGFR mutant NSCLC (99,100). Activation of AXL signaling pathway can occur through overexpression of AXL or its ligand GAS6. Small-molecule AXL inhibitors, MP-470 and XL-880 were able to restore the TKI sensitivity in TKI resistant NSCLC cells. Forced overexpression of AXL in TKI sensitive NSCLC cells can confer TKI resistance. These investigators also found an association between the overexpression of AXL and vimentin, a marker of EMT in the TKI resistant NSCLC cells. In their exploratory analysis of patient samples, approximately 20% of EGFR TKI resistant NSCLC patients were found to have tumors with upregulated AXL, GAS6 and vimentin.
NF-κB activation
NF-κB is an important transcription regulator of the genes that controls cell proliferation and cell growth, including tumor growth. Bivona et al. (101) reported previously that activation of NF-κB signaling pathway can confer TKI resistance in EGFR mutant NSCLC cells. The investigators introduced a shRNA library to target >2,000 cancer relevant genes in the TKI insensitive H1635 NSCLC cell line. This line had an EGFR mutation, but no other identifiable mutations or activation of alternative signaling pathways that could confer insensitivity to EGFR TKI. Among the screen hits conferring TKI sensitivity in H1635, 18 target genes were linked to the NF-κB signaling. Inhibition of NF-κB signaling could enhance TKI sensitivity in H1635 and other EGFR-mutant NSCLC cells, and they reported that higher NF-κB activation state was correlated with worse PFS and decreased overall survival in EGFR-mutant NSCLC patients treated with TKI. However, a recent clinical study of the combination of PF-3512676, an inhibitor for toll-like receptor 9 which activates NF-κB, and erlotinib did not increase PFS as compared to erlotinib alone in patients with advanced recurrent EGFR-mutant NSCLC patients (102).
IGF1-R and KDM5A activation
Sharma et al. (103) reported that a subpopulation of NSCLC tumors developed reversible TKI resistance by engaging the IGF1-R signaling pathway and an altered chromatin state due to a histone demethylase, KDM5A. These TKI resistant cells had upregulated IGFBP-3, KDM5A and increased phosphorylation of IGF-1R. In this subpopulation, IGF1-R inhibitor, depletion of KDM5A or histone deacetylases (HDACs) could markedly suppress the TKI-resistant outgrowth of NSCLC cells in combination with TKI by restoring the TKI sensitivity of TKI resistant cells. Furthermore, inhibition of IGF1-R could lead to decreased KDM5A expression and restoration of H3K4 methylation, suggesting a direct link between IGF-1R signaling pathway and KDM5A function. Altogether, the authors demonstrated that a transient altered chromatin state could potentially mediate TKI resistance in NSCLC. Unfortunately, a recent randomized Phase II study concluded that the combination of IGF1-R inhibitor (R1507) with erlotinib did not provide any PFS or survival advantage over erlotinib alone in unselected NSCLC patients (104). A clinical study to evaluate the combination of erlotinib and HDAC inhibitor, SNDX-275 vs. erlotinib alone in treatment of NSCLC patients has just been completed (NCT000602030), but the results have not been reported.
Other alternative signaling pathway activation
Recently many more signaling pathways have been reported to mediate resistance to EGFR TKI in NSCLC models, but as yet lack evidence for efficacy in patients. These pathways include: activation of Wnt-tankyrase-β-catenin pathway; reduced expression of NF1; downregulation of DAPK through DNA methylation of its CpG island; overexpression of FGF2 and FGFR1 in FGF2-FGFR1 autocrine pathway; upregulation of ADAM17 in heregulin-HER3 autocrine loop; activation of JAK2-related signaling pathway; overexpression of ROR1 caused by NKX2-1; activation of VEGF signaling pathway in stromal cells; overexpression of Notch-1 and its enhancement of EMT; loss of IGF binding proteins; acquisition of stem-cell like properties; and involvement of tumor stroma and cancer-associated fibroblasts derived from EGFR-TKI-resistant tumors (105-118). Many of these pathways have been known to be relevant in cancer development and progression.
Current clinical strategies to overcome AR
When patients relapse secondary to AR, alternative treatment strategies are desired. There is increasing evidence to support patient tumor rebiopsy upon development of resistance to determine the optimal second-line treatments; some cancer centers and clinical trials are already implementing this strategy (119,120). For various cancer sites, rebiopsy is a fairly simple procedure. For lung cancer patients, however, rebiopsy is often a highly invasive procedure, and in many cases, there is a difficult choice of which of multiple metastatic sites should be considered for biopsy. Some patients who develop initial resistance to an EGFR TKI respond again upon a second challenge, after a defined period of a TKI drug holiday (121-124). Song et al. reported that, based on multiple studies, over 50% of patients who progressed on a first line EGFR TKI and then stopped the TKI treatment, benefited from a subsequent second course of the same EGFR TKI (125). There is currently a poor understanding of the mechanisms of reversal of resistance conferred by such a drug holiday.
Optimal therapies have not been established for the majority of EGFR-mutant lung cancer patients who develop disease progression after merely 10 to 14 months on TKIs (20,24,126). Table 1 summarizes the results of clinical trials to date using second and third generations TKIs that were supposed to overcome AR. Second-generation EGFR TKIs have been developed to overcome resistance, however, results from clinical trials have not been as promising as was anticipated.
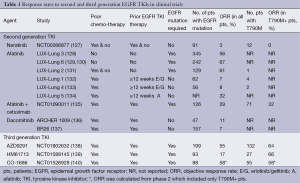
Full table
Second-generation EGFR TKIs form irreversible covalent bonds with the ATP-binding site of EGFR as well as other members of the HER family of receptors (excluding Her3). Neratinib (HK1-272) did not show good response rates (RR) in patients with T790M mutations thus further development was halted (127). Afatinib (BIBW2992) has been investigated as a second- and third-line treatment in patients who have AR to first-generation EGFR TKIs (LUX-Lung 1, 4, and 5 program) and as a first-line treatment in EGFR-mutant patients (LUX-Lung 2, 3, 6 and 7). Thus far, afatinib has been shown to improve the disease control rate and prolong PFS in both LUX-Lung 1 and 2 (131,132). The LUX-Lung 4 trial demonstrated a modest benefit of afatinib as a third- or fourth-line treatment for patients who had previously progressed while receiving erlotinib and/or gefitinib (133). The LUX-Lung 5 trial demonstrated the benefit of combining paclitaxel with afatinib after patients with AR to gefitinib and/or erlotinib progress on afatinib monotherpy (134). Dacomitinib (PF-00299804), another second-generation, irreversible pan-HER TKI, has shown activity against NSCLC cell lines that harbor the T790M mutation. Dacomitinib efficacy was studied in two phase II trials. The first was to evaluate benefit (compared to erlotinib) after failure of one or two chemotherapy regimens, the second compared its benefit as a second- or third-line treatment in patients with advanced NSCLC after failure of at least one prior chemotherapy regimen and prior treatment with erlotinib (141,142). While the results of these two studies seemed initially promising, two randomized phase 3 studies, the ARCHER 1009 trial and the NCIC CTG BR.26 trial, failed to meet their objectives (136,137). The ARCHER 1009 trial did not demonstrate any statistically significant PFS in advanced NSCLC patients treated with dacomitinib compared to erlotinib in the second- and third-line therapy of advanced NSCLC (136). The NCIC CTG BR.26 trial, which included patients with advanced NSCLC who failed previous standard therapy with both chemotherapy and an EGFR TKI, failed to demonstrate significant prolongation of overall survival in those treated with dacomitinib versus placebo, though there was significant improvement in response rate, PFS and time to symptom deterioration in patients with KRAS WT NSCLC (137). In neither of these trials were patients selected specifically for the presence of the T790M mutation.
Recent studies have demonstrated the benefit of combining therapies in overcoming resistance that arises through secondary mutations in the driver oncogene. In both cell line-derived and transgenic mouse models harboring T790M mutations, concurrent administration of the irreversible EGFR TKI, afatinib, and EGFR monoclonal antibody, cetuximab, resulted in dramatic tumor shrinkage (143). A phase I/II trial investigating the same drug combination in NSCLC patients with EGFR mutations and AR to EGFR TKIs demonstrated responses in 40% of patients (135,143). The mechanisms underlying the synergistic effect of this combination appear to be a dramatic inhibition of both phosphorylated EGFR and total EGFR. In contrast, afatinib appears to affect only phosphorylated EGFR and cetuximab appears to only affect the total EGFR protein expression (143). Meador et al. (144) developed resistance to the afatinib/cetuximab combination in PC-9/BRc1- (exon19 deletion/T790M mutant EGFR NSCLC cell line) derived xenografts and found that this occurred via the additional amplification of the EGFR gene. They further demonstrated sensitivity in this resistant model to the third-generation EGFR TKI AZD9291.
Third-generation EGFR TKIs specifically target both activating mutations and T790M mutations in EGFR. These agents seem promising; early results from phase I trials on three 3rd generation EGFR TKIs were presented at the 2014 ASCO Annual Meeting. The first study of HM61713 in advanced NSCLC patients with EGFR mutations who had failed previous EGFR TKIs (NCT01588145) demonstrated disease control rates of 76.5% when treated <4 weeks, and 73.1% when treated ≥4 weeks; 18 of 27 patients carrying T790M mutations showed a decrease in the target lesion sizes (139). The use of AZD9291 in EGFR mutant NSCLC patients (NCT01802632) resulted in (unconfirmed) response rates of 64% in 89 patients with T790M (with disease control in 96%) and only 23% in 43 patients without T790M mutations documented. Importantly, RECIST responses were observed at all dose levels and in brain metastases (138). For the 3rd generation EGFR TKI, CO-1686 (NCT01526928), preliminary results found that, of nine patients carrying T790M mutations, six demonstrated partial responses (PRs), two achieved stable disease, and the final patient achieved PR after transitioning to the HBr form of CO-1686 (140). Despite these promising, early clinical results, resistance to at least one of these third-generation TKIs, CO-1686, has already been demonstrated by an EMT mechanism (145).
Summary
Targeting EGFR in NSCLC patients with activating mutations holds great promise, however AR remains a currently insurmountable hurtle. Mechanisms behind AR have been identified in patients, such as secondary mutations within EGFR, activation of alternate proteins that are downstream of EGFR signaling or activation of proteins that feed into the EGFR signaling cascade. Further mechanisms of AR have been identified in cell lines and remain to be observed in patients. Novel treatment regimens of EGFR TKIs in combination with therapies that target EGFR in different ways or that target alternate proteins are being attempted to overcome known mechanisms of resistance. Third generation EGFR TKIs are being developed in the hopes of overcoming the most common mechanisms of resistance, T790M; to date, the results are preliminary but excitingly optimistic.
Acknowledgements
This work is partially supported by The Rachelle Archambault Innovation Grant of the Canadian Cancer Society (grant #701637) and the Ontario Ministry of Long Term Health. Erin Stewart is supported by the Terry Fox Foundation Strategic Initiative for Excellence in Radiation Research for the 21st Century (EIRR21) at CIHR and the Ontario Graduate Scholarship fund. Dr. Tsao is the M. Qasim Choksi Chair in Lung Cancer Translational Research. Dr. Geoffrey Liu holds the Alan B. Brown Chair of Molecular Genomics and Cancer Care Ontario (CCO) chair in Experimental Therapeutics and Population Studies and is further supported by Poslun family foundation.
Disclosure: Dr. Tsao received honoraria from AstraZeneca, Hoffmann-LaRoche, Boehringer-Ingelheim Canada, Novartis and Pfizer, and research grant from Hoffmann-LaRoche. Dr. Liu received honorarium from AstraZeneca, Hoffmann-LaRoche, Novartis and Pfizer. The other authors declare no conflicts of interest.
References
- Canadian Cancer Statistics publication. Available online: http://www.cancer.ca/en/cancer-information/cancer-101/canadian-cancer-statistics-publication/?region=on
- WHO, Cancer. Available online: http://www.who.int/mediacentre/factsheets/fs297/en/
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [PubMed]
- Chen YM. Update of epidermal growth factor receptor-tyrosine kinase inhibitors in non-small-cell lung cancer. J Chin Med Assoc 2013;76:249-57. [PubMed]
- Kris MG, Johnson BE, Kwiatkowski DJ, et al. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI’s Lung Cancer Mutation Consortium (LCMC). J Clin Oncol 2011;29:abstr CRA7506.
- Shiau CJ, Babwah JP, da Cunha Santos G, et al. Sample features associated with success rates in population-based EGFR mutation testing. J Thorac Oncol 2014;9:947-56. [PubMed]
- Sequist LV, Joshi VA, Jänne PA, et al. Response to treatment and survival of patients with non-small cell lung cancer undergoing somatic EGFR mutation testing. Oncologist 2007;12:90-8. [PubMed]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010;10:760-74. [PubMed]
- Wood ER, Truesdale AT, McDonald OB, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res 2004;64:6652-9. [PubMed]
- Zhang X, Gureasko J, Shen K, et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006;125:1137-49. [PubMed]
- Choi SH, Mendrola JM, Lemmon MA. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene 2007;26:1567-76. [PubMed]
- Shan Y, Eastwood MP, Zhang X, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell 2012;149:860-70. [PubMed]
- Dixit A, Verkhivker GM. Structure-functional prediction and analysis of cancer mutation effects in protein kinases. Comput Math Methods Med 2014;2014:653487.
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [PubMed]
- Stella GM, Luisetti M, Inghilleri S, et al. Targeting EGFR in non-small-cell lung cancer: lessons,experiences, strategies. Respir Med 2012;106:173-83. [PubMed]
- Antonicelli A, Cafarotti S, Indini A, et al. EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int J Med Sci 2013;10:320-30. [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [PubMed]
- Majem M, Remon J. Tumor heterogeneity: evolution through space and time in EGFR mutant non small cell lung cancer patients. Lung Cancer Res 2013;226-37.
- Oxnard GR, Arcila ME, Chmielecki J, et al. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res 2011;17:5530-7. [PubMed]
- Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 2010;277:301-8. [PubMed]
- Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013;8:179-84. [PubMed]
- Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res 2007;67:11924-32. [PubMed]
- Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008;27:4702-11. [PubMed]
- Yuza Y, Glatt KA, Jiang J, et al. Allele-dependent variation in the relative cellular potency of distinct EGFR inhibitors. Cancer Biol Ther 2007;6:661-7. [PubMed]
- Walter AO, Sjin RT, Haringsma HJ, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov 2013;3:1404-15. [PubMed]
- Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31. [PubMed]
- Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res 2008;14:4877-82. [PubMed]
- Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med 2005;2:e313. [PubMed]
- Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013;5:216ra177.
- Tabara K, Kanda R, Sonoda K, et al. Loss of activating EGFR mutant gene contributes to acquired resistance to EGFR tyrosine kinase inhibitors in lung cancer cells. PLoS One 2012;7:e41017. [PubMed]
- Watanabe S, Minegishi Y, Yoshizawa H, et al. Effectiveness of gefitinib against non-small-cell lung cancer with the uncommon EGFR mutations G719X and L861Q. J Thorac Oncol 2014;9:189-94. [PubMed]
- Ohashi K, Maruvka YE, Michor F, et al. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol 2013;31:1070-80. [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [PubMed]
- Godin-Heymann N, Bryant I, Rivera MN, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res 2007;67:7319-26. [PubMed]
- Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res 2007;67:2325-30. [PubMed]
- Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res 2006;66:7854-8. [PubMed]
- Tokumo M, Toyooka S, Ichihara S, et al. Double mutation and gene copy number of EGFR in gefitinib refractory non-small-cell lung cancer. Lung Cancer 2006;53:117-21. [PubMed]
- Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol 2008;26:2442-9. [PubMed]
- Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011;17:1169-80. [PubMed]
- Jänne PA, Borras AM, Kuang Y, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res 2006;12:751-8. [PubMed]
- Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol 2004;26:147-64. [PubMed]
- Querings S, Altmüller J, Ansén S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One 2011;6:e19601. [PubMed]
- Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011;17:1160-8. [PubMed]
- Rosell R, Molina-Vila MA, Taron M, et al. EGFR compound mutants and survival on erlotinib in non-small cell lung cancer (NSCLC) patients (p) in the EURTAC study. J Clin Oncol 2012;30:abstr 7522.
- Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol 2012;7:1640-4. [PubMed]
- Girard N, Lou E, Azzoli CG, et al. Analysis of genetic variants in never-smokers with lung cancer facilitated by an Internet-based blood collection protocol: a preliminary report. Clin Cancer Res 2010;16:755-63. [PubMed]
- Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. [PubMed]
- Ikeda K, Nomori H, Mori T, et al. Novel germline mutation: EGFR V843I in patient with multiple lung adenocarcinomas and family members with lung cancer. Ann Thorac Surg 2008;85:1430-2. [PubMed]
- Ohtsuka K, Ohnishi H, Kurai D, et al. Familial lung adenocarcinoma caused by the EGFR V843I germ-line mutation. J Clin Oncol 2011;29:e191-2. [PubMed]
- Demierre N, Zoete V, Michielin O, et al. A dramatic lung cancer course in a patient with a rare EGFR germline mutation exon 21 V843I: Is EGFR TKI resistance predictable? Lung Cancer 2013;80:81-4. [PubMed]
- Matsushima S, Ohtsuka K, Ohnishi H, et al. V843I, a lung cancer predisposing EGFR mutation, is responsible for resistance to EGFR tyrosine kinase inhibitors. J Thorac Oncol 2014;9:1377-84. [PubMed]
- Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007;4:1669-79; discussion 1680.
- Cragg MS, Kuroda J, Puthalakath H, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med 2007;4:1681-89; discussion 1690.
- Gong Y, Somwar R, Politi K, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med 2007;4:e294. [PubMed]
- Faber AC, Corcoran RB, Ebi H, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov 2011;1:352-65. [PubMed]
- Takezawa K, Okamoto I, Nishio K, et al. Role of ERK-BIM and STAT3-survivin signaling pathways in ALK inhibitor-induced apoptosis in EML4-ALK-positive lung cancer. Clin Cancer Res 2011;17:2140-8. [PubMed]
- Kuribara R, Honda H, Matsui H, et al. Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors. Mol Cell Biol 2004;24:6172-83. [PubMed]
- Kuroda J, Puthalakath H, Cragg MS, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A 2006;103:14907-12. [PubMed]
- Nakagawa T, Takeuchi S, Yamada T, et al. EGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibition. Cancer Res 2013;73:2428-34. [PubMed]
- Costa C, Molina MA, Drozdowskyj A, et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res 2014;20:2001-10. [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [PubMed]
- Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28:S24-31. [PubMed]
- Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med 2011;3:90ra59. [PubMed]
- Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res 2006;12:6494-501. [PubMed]
- Bean J, Riely GJ, Balak M, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res 2008;14:7519-25. [PubMed]
- Costa DB, Schumer ST, Tenen DG, et al. Differential responses to erlotinib in epidermal growth factor receptor (EGFR)-mutated lung cancers with acquired resistance to gefitinib carrying the L747S or T790M secondary mutations. J Clin Oncol 2008;26:1182-4. [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [PubMed]
- Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 2005;97:643-55. [PubMed]
- Yano S, Wang W, Li Q, Matsumoto K, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res 2008;68:9479-87. [PubMed]
- Yamada T, Matsumoto K, Wang W, et al. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin Cancer Res 2010;16:174-83. [PubMed]
- Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. [PubMed]
- Gelsomino F, Facchinetti F, Haspinger ER, et al. Targeting the MET gene for the treatment of non-small-cell lung cancer. Crit Rev Oncol Hematol 2014;89:284-99. [PubMed]
- Xu L, Kikuchi E, Xu C, et al. Combined EGFR/MET or EGFR/HSP90 inhibition is effective in the treatment of lung cancers codriven by mutant EGFR containing T790M and MET. Cancer Res 2012;72:3302-11. [PubMed]
- Ross Camidge D, Ou SH, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32:abstr 8001.
- Zhang YW, Staal B, Essenburg C, et al. Strengthening context-dependent anticancer effects on non-small cell lung carcinoma by inhibition of both MET and EGFR. Mol Cancer Ther 2013;12:1429-41. [PubMed]
- Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922-33. [PubMed]
- Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005;2:e17. [PubMed]
- Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 2012;109:E2127-33. [PubMed]
- Ercan D, Xu C, Yanagita M, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov 2012;2:934-47. [PubMed]
- Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006;116:2695-706. [PubMed]
- Yamasaki F, Johansen MJ, Zhang D, et al. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res 2007;67:5779-88. [PubMed]
- Sos ML, Koker M, Weir BA, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res 2009;69:3256-61. [PubMed]
- Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet 2009;10:336-42. [PubMed]
- Gupta PB, Fillmore CM, Jiang G, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011;146:633-44. [PubMed]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442-54. [PubMed]
- Thomson S, Buck E, Petti F, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res 2005;65:9455-62. [PubMed]
- Rho JK, Choi YJ, Lee JK, et al. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer 2009;63:219-26. [PubMed]
- Suda K, Tomizawa K, Fujii M, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol 2011;6:1152-61. [PubMed]
- Buonato JM, Lazzara MJ. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res 2014;74:309-19. [PubMed]
- Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213-5. [PubMed]
- Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852-60. [PubMed]
- Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. [PubMed]
- Bivona TG, Hieronymus H, Parker J, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature 2011;471:523-6. [PubMed]
- Belani CP, Nemunaitis JJ, Chachoua A, et al. Phase 2 trial of erlotinib with or without PF-3512676 (CPG 7909, a Toll-like receptor 9 agonist) in patients with advanced recurrent EGFR-positive non-small cell lung cancer. Cancer Biol Ther 2013;14:557-63. [PubMed]
- Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69-80. [PubMed]
- Ramalingam SS, Spigel DR, Chen D, et al. Randomized phase II study of erlotinib in combination with placebo or R1507, a monoclonal antibody to insulin-like growth factor-1 receptor, for advanced-stage non-small-cell lung cancer. J Clin Oncol 2011;29:4574-80. [PubMed]
- Casás-Selves M, Kim J, Zhang Z, et al. Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res 2012;72:4154-64. [PubMed]
- de Bruin EC, Cowell C, Warne PH, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov 2014;4:606-19. [PubMed]
- Ogawa T, Liggett TE, Melnikov AA, et al. Methylation of death-associated protein kinase is associated with cetuximab and erlotinib resistance. Cell Cycle 2012;11:1656-63. [PubMed]
- Terai H, Soejima K, Yasuda H, et al. Activation of the FGF2-FGFR1 autocrine pathway: a novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res 2013;11:759-67. [PubMed]
- Ware KE, Hinz TK, Kleczko E, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis 2013;2:e39. [PubMed]
- Ware KE, Marshall ME, Heasley LR, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One 2010;5:e14117. [PubMed]
- Zhou BB, Peyton M, He B, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 2006;10:39-50. [PubMed]
- Harada D, Takigawa N, Ochi N, et al. JAK2-related pathway induces acquired erlotinib resistance in lung cancer cells harboring an epidermal growth factor receptor-activating mutation. Cancer Sci 2012;103:1795-802. [PubMed]
- Yamaguchi T, Yanagisawa K, Sugiyama R, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 2012;21:348-61. [PubMed]
- Naumov GN, Nilsson MB, Cascone T, et al. Combined vascular endothelial growth factor receptor and epidermal growth factor receptor (EGFR) blockade inhibits tumor growth in xenograft models of EGFR inhibitor resistance. Clin Cancer Res 2009;15:3484-94. [PubMed]
- Xie M, Zhang L, He CS, et al. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem 2012;113:1501-13. [PubMed]
- Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest 2008;118:2609-19. [PubMed]
- Shien K, Toyooka S, Yamamoto H, et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell-like properties in cancer cells. Cancer Res 2013;73:3051-61. [PubMed]
- Mink SR, Vashistha S, Zhang W, et al. Cancer-associated fibroblasts derived from EGFR-TKI-resistant tumors reverse EGFR pathway inhibition by EGFR-TKIs. Mol Cancer Res 2010;8:809-20. [PubMed]
- Hata A, Katakami N, Yoshioka H, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer 2013;119:4325-32. [PubMed]
- Stoehlmacher-Williams J, Ehninger G, Zimmermann DR, et al. Targeting TKI-resistance in NSCLC: Importance of rebiopsy and molecular diagnostics—A case study. Cancer Treat Commun 2013;1:1-5.
- Becker A, Crombag L, Heideman DA, et al. Retreatment with erlotinib: Regain of TKI sensitivity following a drug holiday for patients with NSCLC who initially responded to EGFR-TKI treatment. Eur J Cancer 2011;47:2603-6. [PubMed]
- Jang SH. Long Term Therapeutic Plan for Patients with Non-Small Cell Lung Cancer Harboring EGFR Mutation. Tuberc Respir Dis (Seoul) 2014;76:8-14. [PubMed]
- Oxnard GR, Janjigian YY, Arcila ME, et al. Maintained sensitivity to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer recurring after adjuvant erlotinib or gefitinib. Clin Cancer Res 2011;17:6322-8. [PubMed]
- Song Z, Yu X, He C, et al. Re-administration after the failure of gefitinib or erlotinib in patients with advanced non-small cell lung cancer. J Thorac Dis 2013;5:400-5. [PubMed]
- Song T, Yu W, Wu SX. Subsequent treatment choices for patients with acquired resistance to EGFR-TKIs in non-small cell lung cancer: restore after a drug holiday or switch to another EGFR-TKI? Asian Pac J Cancer Prev 2014;15:205-13. [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [PubMed]
- Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol 2010;28:3076-83. [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [PubMed]
- Wu YL, Zhou C, Hu CP, et al. LUX-Lung 6: A randomized, open-label, phase III study of afatinib (A) versus gemcitabine/cisplatin (GC) as first-line treatment for Asian patients (pts) with EGFR mutation-positive (EGFR M+) advanced adenocarcinoma of the lung. J Clin Oncol 2013;31:abstr 8016.
- Yang JC, Shih JY, Su WC, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): a phase 2 trial. Lancet Oncol 2012;13:539-48. [PubMed]
- Miller VA, Hirsh V, Cadranel J, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol 2012;13:528-38. [PubMed]
- Katakami N, Atagi S, Goto K, et al. LUX-Lung 4: a phase II trial of afatinib in patients with advanced non-small-cell lung cancer who progressed during prior treatment with erlotinib, gefitinib, or both. J Clin Oncol 2013;31:3335-41. [PubMed]
- Schuler MH, Yang CH, Park K, et al. Continuation of afatinib beyond progression: Results of a randomized, open-label, phase III trial of afatinib plus paclitaxel (P) versus investigator’s choice chemotherapy (CT) in patients (pts) with metastatic non-small cell lung cancer (NSCLC) progressed on erlotinib/gefitinib (E/G) and afatinib—LUX-Lung 5 (LL5). J Clin Oncol 2014;32:abstr 8019.
- Janjigian YY, Smit EF, Groen HJ, et al. Dual inhibition of EGFR with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov 2014;4:1036-45. [PubMed]
- Ramalingam SS, Janne PA, Mok T, et al. Randomized, double-blinded study of dacomitinib, an irreversible pan-human epidermal growth factor receptor (HER) inhibitor, versus erlotinib for second-line/third-line therapy of locally advanced/metastatic non-small cell lung cancer (ARCHER 1009). J Clin Oncol 2014;32:abstr 7501.
- Ellis PM, Liu G, Millward M, et al. NCIC CTG BR.26: A phase III randomized, double blind, placebo controlled trial of dacomitinib versus placebo in patients with advanced/metastatic non-small cell lung cancer (NSCLC) who received prior chemotherapy and an EGFR TKI. J Clin Oncol 2014;32:abstr 1586.
- Jänne PA, Ramalingam SS, Yang JC, et al. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients (pts) with EGFR inhibitor–resistant non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32:abstr 8010.
- Kim DW, Lee DH, Kang JH, et al. Clinical activity and safety of HM61713, an EGFR-mutant selective inhibitor, in advanced non-small cell lung cancer (NSCLC) patients (pts) with EGFR mutations who had received EGFR tyrosine kinase inhibitors (TKIs). J Clin Oncol 2014;32:abstr 8009.
- Sequist LV, Soria JC, Gadgeel SM, et al. First-in-human evaluation of CO-1686, an irreversible, highly selective tyrosine kinase inhibitor of mutations of EGFR (activating and T790M). J Clin Oncol 2014;32:abstr 8010^.
- Ramalingam SS, Blackhall F, Krzakowski M, et al. Randomized phase II study of dacomitinib (PF-00299804), an irreversible pan-human epidermal growth factor receptor inhibitor, versus erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2012;30:3337-44. [PubMed]
- Janne PA, Reckamp K, Koczywas M, et al. Efficacy and safety of PF-00299804 (PF299) in patients (pt) with advanced NSCLC after failure of at least one prior chemotherapy regimen and prior treatment with erlotinib (E): A two-arm, phase II trial. J Clin Oncol 2009;15:abstr 8063.
- Regales L, Gong Y, Shen R, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest 2009;119:3000-10. [PubMed]
- Meador CB, Jin H, de Stanchina E, et al. Abstract B10: Acquired resistance to afatinib plus cetuximab in EGFR-mutant lung adenocarcinoma may be mediated by EGFR overexpression and overcome by the mutant-specific EGFR inhibitor, AZD9291. Clin Cancer Res 2014;20:B10.
- Walter AO, Sjin RT, Haringsma HJ, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov 2013;3:1404-15. [PubMed]


