Prognostic and predictive biomarkers in early stage non-small cell lung cancer: tumor based approaches including gene signatures
Introduction
Surgery remains the only potentially curative treatment for early-stage non-small cell lung cancer patients (NSCLC) resulting in 5-year survival rates ranging from 77% in pathological stage IA to 23% in stage IIIA tumors (1).
Clinical trials and meta-analyses have demonstrated that in patients with early-stage NSCLC adjuvant chemotherapy improves survival (2-6) with an average benefit of 5% at 5 years and, consequently, adjuvant chemotherapy is recommended for patients with resected stage II-III NSCLC (7-9). Nevertheless, a proportion of stage I patients have poor prognosis and may benefit significantly from adjuvant chemotherapy, while some relatively good prognosis stage II patients may not share similar benefits. Therefore, new diagnostic paradigms are urgently needed to select stage I-II subjects who may take advantage from adjuvant chemotherapy and clinical trials.
The strongest clinical prognostic factors in NSCLC include stage, sex, age, and performance status (10-12), but a better individualization of treatment approaches requires a more precise understanding of the molecular features of lung cancer.
A wide array of individual molecular markers have been tested in advanced as well as early stage NSCLC for prognostic and predictive value and this review will focus on the current existing evidence to support their investigational value. Most of these molecular markers have been found to be either prognostic and/or predictive at the same time. One of the first and largest retrospective biomarker studies in 515 resected stage I NSCLC patients failed to show any significant association between survival and the expression of an extensive panel of biomarkers, including epidermal growth factor receptor (EGFR), HER2/neu, bcl-2, p53 and angiogenesis markers (13).
Lastly, the human genome project, allowed the development and clinical applications of genomic-based assays, including increasingly dense microarray platforms for global analyses of gene expression, copy number variation, DNA methylation and microRNA and several genomic signatures have been identified and tested in early stage NSCLC for their prognostic value.
Excision repair cross-complementation group 1
Cisplatin inhibits replication by binding to DNA and forming platinum-DNA adducts causing strand breaks when the DNA helices unwind in preparation for replication. The nuclear excision repair (NER) family of genes is involved in repair of these DNA strand breaks (14). The excision repair cross-complementation group 1 (ERCC1) enzyme is involved in the final step of the NER pathway that recognizes and removes cisplatin-induced DNA adducts, therefore, leading to cisplatin resistance. High tumoral ERCC1 expression, therefore, predicts for cisplatin resistance.
ERCC1 activity may be assessed as protein by standard immunohistochemistry (IHC) or automated AQUA technology (15). Alternatively, it can be assessed at the mRNA level through quantitative real-time polymerase chain reaction (qRT-PCR). Currently, there is no consensus about the superiority of one approach versus the other because both techniques are rarely assessed concomitantly (16,17).
In resected NSCLC, patients with high ERCC1 expression (>50 unitless ratio) had a better survival outcome (median OS, 94.6 vs. 35.5 months; P=0.01) when compared to patients with low ERCC1 expression generating the hypothesis that an intact DNA repair mechanism may reduce the accumulation of genetic aberrations that are thought to contribute to malignant potential phenotype and therefore the risk of relapse after definitive treatment (18).
The predictive role of ERCC1 was initially assessed by RT-PCR in a series of small retrospective studies in advanced NSCLC (19,20). The median overall survival (OS) was significantly longer in patients with low ERCC1 compared to patients with high ERCC1. Subsequently, ERCC1 was investigated in a subgroup of patients enrolled in a large adjuvant chemotherapy trial (21) and, by standard immunohistochemistry, in 761 paraffin-embedded tumor samples (22). A benefit from cisplatin-based adjuvant chemotherapy was associated with the absence or low expression of ERCC1 (test for interaction, P=0.009) with a significantly prolonged disease-free survival and OS among patients with ERCC1-negative tumors (HR for death, 0.65; 95% CI, 0.50-0.86; P=0.002) as opposed to ERCC-1 positive tumors. Morever, the prognostic value of ERCC1 was confirmed in the control group with a significantly higher 5-year OS among patients with ERCC1-positive tumors than negative ones (HR, 0.66; 95% CI, 0.49-0.90; P=0.009). The same tumors were also scored by AQUA and low ERCC1 scores were marginally prognostic (HR =0.77 for high versus low scores, P=0.10) (23) while in an additional study, ERCC1 was exclusively predictive in squamous cell carcinoma (24).
The specificity of the commonly used mouse monoclonal antibody 8F1 to ERCC1 has been extensively debated (25,26). It was also found that none of the 16 antibodies tested could distinguish among the four known ERCC1 protein isoforms (27). In the neo-adjuvant setting, mRNA ERCC1 levels in pretreatment tissue samples were correlated with the capacity to achieve an objective response following platinum-based chemotherapy (P<0.05), but not with the formation of local or distant metastases (28).
The predictive value of ERCC1 was enhanced by the concurrent evaluation of MutS homolog 2 (MSH2), a major active component of the mismatch repair system. Patients with double-negative tumors experienced a greater benefit from chemotherapy (29).
Overall these data indicate a prognostic role of ERCC1 expression while its predictive role remains to be further assessed and additional validation studies are needed.
Breast cancer susceptibility gene 1
The protein encoded by breast cancer susceptibility gene 1 (BRCA1) has a crucial role in DNA repair as well as in cell-cycle checkpoints and mitotic spindle assembly (30). BRCA1 sensitizes cancer cells to apoptosis induced by antimicrotubule drugs, such as taxanes and vinca alkaloids, while conferring resistance to DNA-damaging agents, including platinum agents. The potential prognostic role of a panel of nine candidate biomarkers including BRCA1 was investigated in two independent cohorts of chemotherapy naive patients with early-stage NSCLC. BRCA1 was the only independent factor affecting OS (31). In a group of patients with locally advanced NSCLC, treated with neo-adjuvant cisplatin and gemcitabine followed by surgery, those with the lowest levels of BRCA1 mRNA expression had significantly greater benefit from chemotherapy in terms of clinical and pathological downsizing and OS (32).
For its localization to sites of DNA double strand breaks, the upstream activity of the receptor-associated protein 80 (RAP-80) is required for BRCA1. In a first line study in advanced NSCLC patients with the lowest expression of both BRCA1 and RAP-80 receiving cisplatin plus gemcitabine it was shown that RAP-80 can modulate the effect of BRCA1. In addition to a close correlation with BRCA1, RAP-80 expression was identified as an independent predictor for OS (33). In a phase II feasibility study of adjuvant chemotherapy in patients with stage II-IIIA NSCLC, those with high BRCA1 transcriptional levels received single agent docetaxel, whereas those with intermediate and low BRCA1 expression were treated with cisplatin-doublets and OS did not differ between the treatment arms (34).
A recent meta-analysis of 23 studies assessed the role of BRCA1 as a predictor of clinical outcome in platinum- and paclitaxel-based chemotherapy in NSCLC patients. In 17 platinum-based studies, low/negative BRCA1 was associated with better objective response rate [ORR] (OR =1.70, 95% CI, 1.32-2.18), longer OS and event-free survival [EFS] (HR =1.58, 95% CI, 1.27-1.97, and HR =1.60, 95% CI, 1.07-2.39 for OS and EFS, respectively). In 4 paclitaxel-based chemotherapy studies, patients with high/positive BRCA1 had better ORR (OR =0.41, 95% CI, 0.26-0.64) while OS and EFS were not evaluated because of the insufficient data available (35). Some studies reported that ERCC1 expression is closely linked to RRM1 and BRCA1 levels (32,36,37), with concordant levels in 70-80% of cases (20,38).
Ribonucleotide reductase M1
Ribonucleotide reductase M1 (RRM1) is the regulatory component of an essential enzyme that catalyzes the reduction of ribonucleoside diphosphates to the corresponding deoxyribonucleotides. A role for ribonucleotide reductase in DNA repair has been proposed, given the capacity of RRM1 to bind a p53-regulated paralog of RRM2 called p53R2 (39,40). Increased RRM1 predicts for decreased tumor invasiveness and metastatic potential, therefore predicting for more indolent behavior, perhaps mediated through its direct correlation with phosphatase and PTEN (phosphatase and tensin homolog) protein expression (41,42). In resected NSCLC, RRM1 protein proved prognostic, with low levels associated with a median OS of 60.2 months, compared to more than 120 months in high RRM1 tumors (43). RRM1 is a major predictor of disease response to gemcitabine, being its predominant target, as well as platinum (44). Several studies investigated the predictive value of RRM1 in patients treated with gemcitabine plus cisplatin (45,46) demonstrating that RRM1 expression was significantly and inversely correlated with disease response, though not with survival (47). A recent meta-analysis in 1,243 patients with advanced NSCLC treated with gemcitabine-based regimens concluded that low tumor RRM1 was associated with a better response rate and longer survival (48).
Thymidylate synthase
Thymidylate synthase (TS) catalyzes the conversion of deoxyuridine monophosphate (dUMP) to (deoxy) thymidine monophosphate (TMP), which requires oxidization of tetrahydrofolate to dihydrofolate. High tumoral levels of TS have been associated with resistance to 5-FU (49-51). Retrospective and prospective data from phase III trials in advanced NSCLC have established the favorable predictive value of TS in non-squamous NSCLC treated with pemetrexed with mRNA and protein TS expression lower in non-squamous compared to squamous histology and small cell lung cancer (52,53).These findings have been recently confirmed in a large retrospective study, although there was a wide range between individual patients (54).
Similarly distinct TS expression patterns among NSCLC subtypes were observed in stage I-IIIA NSCLC (55). In two different cohorts of chemotherapy naive patients with resected early-stage NSCLC, TS was a prognostic factor with mixed results. In one study high TS mRNA (but not protein) expression, was significantly associated with adverse disease free survival (DFS) and in the other study, high TS expression as determined by AQUA but not by qRT-PCR, predicted improved OS (55,56).
ITACA (International TAilored Chemotherapy Adjuvant) (57) trial is a randomized phase III trial comparing adjuvant pharmacogenomic-driven chemotherapy based on ERCC-1 and TS assessment by qRT-PCR versus standard adjuvant chemotherapy in completely resected stage II-IIIA NSCLC. The molecular assessment groups patients into four different genetic profiles with patients dichotomized by high versus low expression of both ERCC-1 and TS. Within 30 to 45 days post-surgery, patients in each genetic profile are randomized to receive either a standard adjuvant chemotherapy doublet (control arm) or an experimental treatment guided by molecular determinants (tailored chemotherapy arm). The study is currently accruing patients in Italy and in Germany and more than 600 patients have been already randomized (see Figure 1).
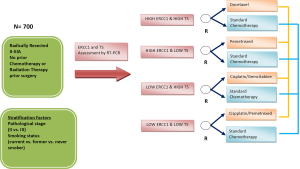
Cyclin-dependent kinase inhibitor 1B
The cyclin-dependent kinase inhibitor 1B, p27Kip1, is a tumor-suppressor protein that induces cell-cycle arrest in phase G1. Despite its anti-proliferative properties, p27Kip1 up-regulation leads to de novo resistance to platinum agents by allowing cancer cells to repair DNA damage and avoid apoptosis. A survival benefit from cisplatin-based chemotherapy was only demonstrated in patients with p27Kip1 negative tumors (58,59). Cyclin D2 has been associated with poor recurrence-free survival in patients in stage III NSCLC treated with surgery with or without adjuvant chemotherapy (60).
β-tubulin
β-tubulin (βTubIII) is an essential element of microtubules, which serves as a cellular structural component involved in vital processes, including mitosis. Class IIIβ-tubulin (βTUBIII) corresponds to an isotype with enhancing impact on microtubule dynamics, contributing to intrinsic cancer cell resistance to antimitotic agents. Several studies have shown that βTubIII expression may predict response and outcome in patients with advanced NSCLC receiving tubulin binding agents (61,62). In patients with early stage NSCLC treated with an adjuvant vinorelbine-based regimen (2), high βTUBIII expression was shown to be an independent adverse predictor of recurrence-free survival (63). The prognostic value was confirmed retrospectively in patients enrolled in another adjuvant study (64) and more recently in a neo-adjuvant study (65). In patients with β-tubulin positive immunostaining, median PFS was 30.6 versus 60.1 months (HR =1.46; 95% CI, 1.08-1.99) for those negative.
RAS oncogene and p53
KRAS mutation was associated in some studies with a poor prognosis in early NSCLC patients (66) but not in others. In the phase III NCIC JBR.10 adjuvant trial, where patients with stage IB-II disease were prospectively stratified by the presence of mutation in any of the RAS genes, the effect of RAS mutation status on the treatment outcome and prognosis was not significant. Nevertheless, the lack of benefit from the cisplatin/vinorelbine combination in patients with RAS mutations, in contrast to the total study population, may suggest a possible negative predictive role (67). Similarly, another adjuvant study in stage IB patients showed that, the presence KRAS mutations trended for an inferior survival of patients with tumors larger than 4 cm (68). Consistent data were reported in an Italian adjuvant study where in a subgroup of 227 patients the presence of K-RAS mutation, but not p53 and Ki67, in the univariate, but not in the multivariate analysis, was associated with shorter survival (69).
p53 nuclear immunoreactivity is considered a surrogate marker of TP53 gene mutations. However, the sensitivity and positive predictive value of p53 IHC expression for TP53 mutation status are estimated to be only 75% and 65%, respectively (70). A retrospective analysis of the phase III NCIC-JBR.10 adjuvant trial showed p53 IHC overexpression to be an independent unfavorable prognostic factor among patients in the observation arm. In contrast to p53 expression, TP53 mutation status was neither prognostic for survival, nor predictive for efficacy of adjuvant chemotherapy (71).
Gene expression signatures
Microarray technologies allow exploration of the prognostic significance of thousands of markers using high-throughput and computational approaches. To date, in lung cancer more than 30 studies have been reported (72) a large number showing that gene expression signature may stratify early stage NSCLC patients with different prognosis or survival outcome.
Although most of these signatures have been validated in one or more independent patient cohorts, microarray dataset overlaps between the genes sets have consistently been minimal. Thus there is a strong possibility that sample collection methods, processing protocols, single-institution subject cohorts, small sample sizes, and peculiarities of the different microarray platforms are contributing significantly to the results. To address these issues, a multi-institutional collaborative study was conducted to generate gene expression profiles from a large number of samples with a priori determined clinical features, useful to evaluate proposed prognostic models for potential clinical implementation. A large series of lung adenocarcinomas were tested for whether microarray measurements of gene expression either alone or combined with basic clinical covariates (stage, age, sex) can be used to predict overall survival in lung cancer subjects. Risk scores were produced substantially correlated with actual subject outcome, especially when clinical and molecular information are combined to build prognostic models for early stage lung cancer (73).
A malignancy-risk gene signature composed of several genes associated with proliferative activity has been successfully applied to predict breast cancer risk (74,75) and also tested for prognostic and predictive value in an early-stage NSCLC patients. The malignancy-risk gene signature was tested using a large NSCLC microarray dataset from the Director’s Challenge Consortium (n=442) and two independent NSCLC microarray datasets (n=117 and 133 datasets, respectively). The malignancy-risk gene signature was significantly associated with OS (P<0.001) in two independent datasets and also with adjuvant chemotherapy (P=0.02) (76). Xu et al. developed an empirical model which is not based on the knowledge of patients’ survival time for determining the lung cancer biomarker signature. It has been hypothesized that instead of an individual gene, two functionally imbalanced groups of genes (Yin and Yang) determine the fate of the tumor cells, which ultimately determines patient’s survival time. The Yin and Yang genes were selected by comparing expression data from normal lung and lung cancer tissue samples using both unsupervised clustering and pathways analyses. The model was tested in four independent lung cancer datasets and significantly stratified patients into high- and low-risk survival groups and predicted chemotherapy outcomes for stages II and III (77).
A 14-gene expression assay that uses quantitative PCR, runs on formalin-fixed paraffin-embedded tissue samples, and differentiates patients with heterogeneous statistical prognoses was developed in a cohort of 361 resected patients with non-squamous NSCLC and validated in 2 different cohorts of 433 patients with resected stage I non-squamous NSCLC and 1,006 patients with stage I-III non-squamous NSCLC resected in several leading Chinese cancer centres. The signature significantly segregated patients in low-, intermediate- and high-risk patients with relevant differences in 5-year survival rate. Multivariate analysis in both cohorts indicated that no standard clinical risk factors could account for, or provide, the prognostic information derived from tumour gene expression. This quantitative-PCR-based assay reliably identified patients with early-stage non-squamous NSCLC at high risk for mortality after surgical resection (78).
Other biomarkers in early NSCLC
Insulin receptor (IR) and Insulin-like growth factor receptor (IGF1R) are implicated in the development and progression of NSCLC, either by interacting with the EGFR pathway or independently. In patients with resected NSCLC, IGF1R amplification determined by FISH was an independent favorable prognostic factor, unlike IGF1R protein (79). It has been observed that in early stage NSCLC, IGF1R/EGFR FISH+ and IGF1R/EGFR IHC+ were associated with shorter disease-free survival (P=0.05 and P=0.05, respectively). Patients with concomitant IGF1R/EGFR FISH+/IHC+ had a worse DFS and OS (P=0.005 and P=0.01, respectively) (80).
Hepatocyte growth factor receptor (c-MET) is a proto-oncogene associated with tumor invasive growth. In patients with resected stage I-III carcinomas, not treated with adjuvant chemotherapy, a high MET gene copy number was an independent adverse prognostic factor mainly in squamous histotype (81). Similarly, in patients with early-stage NSCLC HER2 expression was associated with poorer prognosis especially in stages IB and IIA diseases (82).
Higher expression of CXCR7 is associated with metastatic progression and poor DFS in patients with stage I NSCLC (83). In a retrospective analysis of completely resected stage I tumors, patients with CXCR4-positive tumors had a significantly longer survival than patients with CXCR4-negative tumors (P=0.039). Interestingly, the 5-year metastasis rates were 23.5% and 34.1% in patients with CXCR4-positive and CXCR4-negative expression, respectively (P=0.2) (84).
Recently, a set of genes with altered methylation status were identified in stage I NSCLCs, some of which associated with survival. Such newly identified potential candidates for a NSCLC molecular screening need further analysis in order to determine their clinical utility (85,86).
Studies in early stage NSCLC have reported an association between vascular endothelial growth factor (VEGF) over-expression and progression or poor survival (87,88), but overall the prognostic role of VEGF expression in NSCLC remains undetermined.
Conclusions
Currently, tumor stage remains the strongest predictor of survival in NSCLC. Early-stage patients are treated primarily by surgical resection. However, 30% to 55% of these patients develop recurrence and die of their disease. The current staging system is inadequate for predicting the outcome of treatment and the prognosis in individual patients. For this reason there is an urgent need to search for new individual biomarkers and gene signatures in the tumor tissue. Many studies have investigated several molecular alterations in early lung cancer and their predictive and prognostic implications are still a matter of clinical research and even for the most promising are not yet ready for prime time application. Currently, randomized clinical trials are exploring the real value of these new diagnostic tools.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. [PubMed]
- Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. [PubMed]
- Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol 2006;7:719-27. [PubMed]
- Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004;350:351-60. [PubMed]
- Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. [PubMed]
- NSCLC Meta-analyses Collaborative Group, Arriagada R, Auperin A, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet 2010;375:1267-77. [PubMed]
- NCCN Clinical practice guidelines in oncologyTM. Non Small Cell Lung Cancer.V.1.2011. Available online: http://www.nccn.org/professionals/physician gls/PDF.nscl.pdf
- Vansteenkiste J, De Ruysscher D, Eberhardt WEE, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;00:1-10.
- Pisters KM, Evans WK, Azzoli CG, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I-IIIA resectable non small-cell lung cancer guideline. J Clin Oncol 2007;25:5506-18. [PubMed]
- Lau CL, D’Amico DA, Harpole DH Jr. Clinical and molecular prognostic factors and models for non-small cell lung cancer. In: Pass HI, Mitchell JB, Johnson DH, et al. eds. Lung Cancer Principles and Practice (ed 2). Philadelphia, PA, Lippincott Williams & Wilkins 2000:602-11.
- Graziano SL. Non-small cell lung cancer: clinical value of new biological predictors. Lung Cancer 1997;17:S37-S58. [PubMed]
- Takise A, Kodama T, Shimosato Y, et al. Histopathologic prognostic factors in adenocarcinomas of the peripheral lung less than 2 cm in diameter. Cancer 1988;61:2083-8. [PubMed]
- Pastorino U, Andreola S, Tagliabue E, et al. Immunocytochemical markers in stage I lung cancer: relevance to prognosis. J Clin Oncol 1997;15:2858-65. [PubMed]
- Altaha R, Liang X, Yu JJ, et al. Excision repair cross complementing-group 1: gene expression and platinum resistance. Int J Mol Med 2004;14:959-70. [PubMed]
- Metro G, Zheng Z, Fabi A, et al. In situ protein expression of RRM1, ERCC1, and BRCA1 in metastatic breast cancer patients treated with gemcitabine-based chemotherapy. Cancer Invest 2010;28:172-80. [PubMed]
- Doll CM, Prystajecky M, Eliasziw M, et al. Low ERCC1 mRNA and protein expression are associated with worse survival in cervical cancer patients treated with radiation alone. Radiother Oncol 2010;97:352-9. [PubMed]
- De Castro G Jr, Pasini FS, Siqueira SA, et al. ERCC1 protein, mRNA expression and T19007C polymorphism as prognostic markers in head and neck squamous cell carcinoma patients treated with surgery and adjuvant cisplatin-based chemoradiation. Oncol Rep 2011;25:693-9. [PubMed]
- Simon GR, Sharma S, Cantor A, et al. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 2005;127:978-83. [PubMed]
- Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 2002;8:2286-91. [PubMed]
- Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol 2006;17:1818-25. [PubMed]
- Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol 2010;28:35-42. [PubMed]
- Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [PubMed]
- Bepler G, Olaussen KA, Vataire AL, et al. ERCC1 and RRM1 in the international adjuvant lung trial by automated quantitative in situ analysis. Am J Pathol 2011;178:69-78. [PubMed]
- Pierceall WE, Olaussen KA, Rousseau V, et al. Cisplatin benefit is predicted by immunohistochemical analysis of DNA repair proteins in squamous cell carcinoma but not adenocarcinoma: theranostic modeling by NSCLC constituent histological subclasses. Ann Oncol 2012;23:2245-52. [PubMed]
- Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. N Engl J Med 2007;356:2538-40. [PubMed]
- Ma D, Baruch D, Shu Y, et al. Using protein microarray technology to screen anti-ERCC1 monoclonal antibodies for specificity and applications in pathology. BMC Biotechnol 2012;12:88. [PubMed]
- Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 2013;368:1101-10. [PubMed]
- Marra A, Kemming D, Krueer, et al. ERCC1 as a predictor of response to induction therapy for stage III non-small cell lung cancer. J Clin Oncol 2013;31:abstr e18511.
- Kamal NS, Soria JC, Mendiboure J, et al. MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clin Cancer Res 2010;16:1206-15. [PubMed]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 2006;34:1416-26. Print 2006.
- Rosell R, Skrzypski M, Jassem E, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One 2007;2:e1129. [PubMed]
- Taron M, Rosell R, Felip E, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet 2004;13:2443-9. [PubMed]
- Rosell R, Perez-Roca L, Sanchez JJ, et al. Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One 2009;4:e5133. [PubMed]
- Cobo M, Massuti B, Moran T, et al. Spanish customized adjuvant trial (SCAT) based on BRCA1 mRNA levels. J Clin Oncol 2008;26;abstr 7533.
- Yang Y, Xie Y, Xian L. Breast cancer susceptibility gene 1 (BRCA1) predict clinical outcome in platinum- and toxal-based chemotherapy in non-small-cell lung cancer (NSCLC) patients: a system review and meta-analysis. J Exp Clin Cancer Res 2013;32:15. [PubMed]
- Rosell R, Felip E, Taron M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res 2004;10:4215s-4219s. [PubMed]
- Zhang GB, Chen J, Wang LR, et al. RRM1 and ERCC1 expression in peripheral blood versus tumor tissue in gemcitabine/carboplatin-treated advanced non-small cell lung cancer. Cancer Chemother Pharmacol 2012;69:1277-87. [PubMed]
- Gandara DR, Grimminger PP, Mack PC, et al. Histology- and gender-related associations of ERCC1, RRM1, and TS biomarkers in 1,802 patients with NSCLC: Implications for therapy. J Clin Oncol 2010;28:7513.
- Smith P, Zhou B, Ho N, et al. 2.6 A X-ray crystal structure of human p53R2, a p53-inducible ribonucleotide reductase. Biochemistry 2009;48:11134-41. [PubMed]
- Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res 2004;10:1318-25. [PubMed]
- Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene 2003;22:2135-42. [PubMed]
- Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol 2004;22:1878-85. [PubMed]
- Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med 2007;356:800-8. [PubMed]
- Fairman JW, Wijerathna SR, Ahmad MF, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol 2011;18:316-22. [PubMed]
- Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol 2006;24:4731-7. [PubMed]
- Bepler G, Sommers KE, Cantor A, et al. Clinical efficacy and predictive molecular markers of neoadjuvant gemcitabine and pemetrexed in resectable non-small cell lung cancer. J Thorac Oncol 2008;3:1112-8. [PubMed]
- Reynolds C, Obasaju C, Schell MJ, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol 2009;27:5808-15. [PubMed]
- Gong W, Zhang X, Wu J, et al. RRM1 expression and clinical outcome of gemcitabine-containing chemotherapy for advanced non-small-cell lung cancer: a meta-analysis. Lung Cancer 2012;75:374-80. [PubMed]
- Johnston PG, Lenz HJ, Leichman CG, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res 1995;55:1407-12. [PubMed]
- Leichman CG, Lenz HJ, Leichman L, et al. Quantification of intratumoral thymidylate synthase expression predicts for disseminated colorectal cancer response and resistance to protracted-infusion fluorouracil and weekly leucovorin. J Clin Oncol 1997;15:3223-3229. [PubMed]
- Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol 2001;19:4298-304. [PubMed]
- Scagliotti G, Hanna N, Fossella F, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two Phase III studies. Oncologist 2009;14:253-63. [PubMed]
- Ceppi P, Volante M, Saviozzi S, et al. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase. Cancer 2006;107:1589-96. [PubMed]
- Maus MK, Mack PC, Astrow SH, et al. Histology-related associations of ERCC1, RRM1, and TS biomarkers in patients with non-small-cell lung cancer: implications for therapy. J Thorac Oncol 2013;8:582-6. [PubMed]
- Shintani Y, Ohta M, Hirabayashi H, et al. New prognostic indicator for non-small-cell lung cancer, quantitation of thymidylate synthase by real-time reverse transcription polymerase chain reaction. Int J Cancer 2003;104:790-5. [PubMed]
- Zheng Z, Li X, Schell MJ, et al. Thymidylate synthase in situ protein expression and survival in stage I nonsmall-cell lung cancer. Cancer 2008;112:2765-73. [PubMed]
- Novello S, Manegold C, Grohe C, et al. International tailored chemotherapy adjuvant trial: Itaca trial. J Clin Oncol 2012;30:abstr TPS7109.
- Rekhtman N, Azzoli CG, Kris MG, et al. Patterns of co-expression of ERCC1 and P27 in resected non-small cell lung cancer by immunohistochemistry. J Clin Oncol 2008;26;abstr 7595.
- Filipits M, Pirker R, Dunant A, et al. Cell cycle regulators and outcome of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer: the International Adjuvant Lung Cancer Trial Biologic Program. J Clin Oncol 2007;25:2735-40. [PubMed]
- Ko E, Kim Y, Park SE, et al. Reduced expression of cyclin D2 is associated with poor recurrence-free survival independent of cyclin D1 in stage III non-small cell lung cancer. Lung Cancer 2012;77:401-6. [PubMed]
- Dumontet C, Isaac S, Souquet PJ, et al. Expression of class III beta tubulin in non-small cell lung cancer is correlated with resistance to taxane chemotherapy. Bull Cancer 2005;92:E25-30. [PubMed]
- Sève P, Mackey J, Isaac S, et al. Class III beta-tubulin expression in tumor cells predicts response and outcome in patients with non-small cell lung cancer receiving paclitaxel. Mol Cancer Ther 2005;4:2001-7. [PubMed]
- Sève P, Lai R, Ding K, et al. Class III beta-tubulin expression and benefit from adjuvant cisplatin/vinorelbine chemotherapy in operable non-small cell lung cancer: analysis of NCIC JBR.10. Clin Cancer Res 2007;13:994-9. [PubMed]
- Reiman T, Seve P, Vataire A, et al. Prognostic value of class III B-tubulin (Tubb3) in operable non-small cell lung cancer (NSCLC) and predictive value for adjuvant cisplatin-based chemotherapy (CT): A validation study on three randomized trials. J Clin Oncol 2008;26;abstr 7506.
- Zalcman G, Levallet G, Bergot E, et al. Evaluation of class III beta-tubulin (bTubIII) expression as a prognostic marker in patients with resectable non-small cell lung cancer (NSCLC) treated by perioperative chemotherapy (CT) in the phase III trial IFCT-0002. J Clin Oncol 2009;27:15;abstr 7526.
- Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol 2009;4:22-9. [PubMed]
- Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol 2007;25:5240-7. [PubMed]
- Capelletti M, Wang XF, Gu L, et al. Impact of KRAS mutations on adjuvant carboplatin/paclitaxel in surgically resected stage IBm NSCLC: CALGB 9633. J Clin Oncol 2010;28; abstr 7008.
- Scagliotti GV, Fossati R, Torri V, et al. Randomized study of adjuvant chemotherapy for completely resected stage I, II, or IIIA non-small-cell Lung cancer. J Natl Cancer Inst 2003;95:1453-61. [PubMed]
- Greenblatt MS, Bennett WP, Hollstein M, et al. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 1994;54:4855-78. [PubMed]
- Schiller JH, Adak S, Feins RH, et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol 2001;19:448-57. [PubMed]
- Zhu CQ, Pintilie M, John T, et al. Understanding prognostic gene expression signatures in lung cancer. Clin Lung Cancer 2009;10:331-40. [PubMed]
- Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma, Shedden K, Taylor JM, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [PubMed]
- Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med 2007;356:217-26. [PubMed]
- Wan YW, Qian Y, Rathnagiriswaran S, et al. A breast cancer prognostic signature predicts clinical outcomes in multiple tumor types. Oncol Rep 2010;24:489-94. [PubMed]
- Chen DT, Hsu YL, Fulp WJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst 2011;103:1859-70. [PubMed]
- Xu W, Banerji S, Davie JR, et al. Yin Yang gene expression ratio signature for lung cancer prognosis. PLoS One 2013;8:e68742. [PubMed]
- Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet 2012;379:823-32. [PubMed]
- Dziadziuszko R, Merrick DT, Witta SE, et al. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol 2010;28:2174-80. [PubMed]
- Ludovini V, Flacco A, Bianconi F, et al. Concomitant high gene copy number and protein overexpression of IGF1R and EGFR negatively affect disease-free survival of surgically resected non-small-cell-lung cancer patients. Cancer Chemother Pharmacol 2013;71:671-80. [PubMed]
- Go H, Jeon YK, Park HJ, et al. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol 2010;5:305-13. [PubMed]
- Xia Q, Zhu Z, Wang J, et al. Expression and association of HER2 with prognosis in early-stage (T1-T2N0M0) non-small cell lung cancer. Tumour Biol 2012;33:1719-25. [PubMed]
- Iwakiri S, Mino N, Takahashi T, et al. Higher expression of chemokine receptor CXCR7 is linked to early and metastatic recurrence in pathological stage I nonsmall cell lung cancer. Cancer 2009;115:2580-93. [PubMed]
- Spano JP, Andre F, Morat L, et al. Chemokine receptor CXCR4 and early-stage non-small cell lung cancer: pattern of expression and correlation with outcome. Ann Oncol 2004;15:613-7. [PubMed]
- Lokk K, Vooder T, Kolde R, et al. Methylation markers of early-stage non-small cell lung cancer. PLoS One 2012;7:e39813. [PubMed]
- de Fraipont F, Levallet G, Creveuil C, et al. An apoptosis methylation prognostic signature for early lung cancer in the IFCT-0002 trial. Clin Cancer Res 2012;18:2976-86. [PubMed]
- Liao M, Wang H, Lin Z, et al. Vascular endothelial growth factor and other biological predictors related to the postoperative survival rate on non-small cell lung cancer. Lung Cancer 2001;33:125-32. [PubMed]
- Baillie R, Carlile J, Pendleton N, et al. Prognostic value of vascularity and vascular endothelial growth factor expression in non-small cell lung cancer. J Clin Pathol 2001;54:116-20. [PubMed]


