Are we ready to use biomarkers for staging, prognosis and treatment selection in early-stage non-small-cell lung cancer?
Introduction
Approximately 25-30% of patients with non-small cell lung cancer (NSCLC) present with early stage disease and undergo surgery with curative intent. Despite complete tumor resection, many of these patients will develop systemic relapses with or without local relapses and will eventually die. A meta-analysis of early trials indicated a trend towards improved survival for adjuvant platinum-based chemotherapy and led to a re-evaluation of adjuvant treatment in clinical trials in large patient populations. Several of these trials demonstrated an improved survival with the use of adjuvant chemotherapy. The survival benefit was then further confirmed in a meta-analysis that included all five cisplatin-based trials (Table 1) (1-7). Early NSCLC comprises a heterogeneous group of diseases, with diverse innate aggressiveness and degree of response to cytotoxic agents. For instance, some patient subsets with stage II or even stage IIIA have excellent prognosis and could be spared the toxicity of unnecessary therapy. Others, such as the elderly or less fit, as well as those with stage I disease, remain undertreated despite potential benefit from adjuvant chemotherapy. This underscores the need for a customized approach to tailor adjuvant chemotherapy according to patient characteristics and tumor characteristics. Personalizing therapy based on an individual patient’s molecular profile is a potentially promising approach to optimize efficacy with available agents. Prognostic biomarkers indicate the natural course of disease, irrespective of treatment, while those defined as predictive can foresee differential therapeutic outcomes. However, some biomarkers combine both of these functions, such as ERCC1 and RRM1. Identification and application of the appropriate biomarkers would enable selection of only high-risk patients to receive the most effective treatment. In this review we describe potential predictive and prognos-tic markers and their current role, benefit, and possible future use in the management of patients with early stage NSCLC.

Full Table
Clinicopathological prognostic and predictive factors in early stage NSCLC
It has long been recognized that differences in clinical factors such as stage, sex, and tumor factors such as cellular differentiation, vascularity, and vascular invasion, are prognostic of outcome and important in determining adjuvant therapy decisions for early stage NSCLC. To date, pathological stage, as defined by tumor size and nodal status, is the only prospectively validated clinicopathological biomarker with both prognostic and predictive value. According to guidelines from the European Society of Medical Oncology (ESMO), American Society of Clinical Oncology (ASCO), National Comprehensive Cancer Network (NCCN) and the American College of Chest Physicians (ACCP), platinum-based adjuvant chemotherapy is considered standard treatment for resected stage II-IIIA disease with an estimated survival benefit of 4-5% at 5 years (8-11). Although pre hoc subgroup analyses have shown no benefit for patients with pathological stage IB disease, interpretation of these results should be cautious, since the test for interaction between treatment effect and stage was not significant in any of the phase III trials with platinum-based regimens (2-4,12). Only one of the available meta-analyses did demonstrate a significant differential treatment effect, largely in the stage IA subgroup, suggesting that patients with stages II and IIIA have greatest benefit (2). The reported 11% 5 years survival gain of adjuvant tegafur-uracil in stage I NSCLC cannot be directly extended to Western countries where tegafur-uracil has not yet been reliably tested (13). At the same time, the assumption of therapeutic benefit for stage IB disease with tumor size larger than 4 cm is based on an unplanned subgroup analysis (5). Nonetheless, worse prognosis observed with increasing T size has been recognized in the 7th TNM edition. T2 was divided into T2a (3-5 cm) and T2b (5-7 cm), with 5 year overall survival of 58% and 49%, respectively (P<0.0001); T2bN0 was upstaged to stage IIA. Correlation with the new staging system failed to validate the 5 cm cut-off in the 9-year update of CALGB 9633, showing a trend towards a significant benefit for adjuvant treatment for patients with tumors >7 cm (HR=0.53; P=0.051), although this interaction should be investigated further (14-16).
With regard to the importance of accurate staging, a recent retrospective analysis of lymph node dissection in more than 20,000 patients with pathological stage I NSCLC implied that the number of recovered lymph nodes might be predictive for survival outcomes, although this could well be attributed to a direct therapeutic effect (17). Furthermore, a large retrospective analysis from the SEER database showed that the increasing number of resected positive nodes and a higher ratio between metastatic and overall resected nodes has an independent negative prognostic impact for overall survival in N1 patients (18,19). Recent studies investigated high tumor grade, vascular invasion and visceral pleural infiltration as poor prognostic determinants, based on mostly retrospective cohort studies, and are commonly recommended as adjunct selection criteria for patients who are borderline candidates for adjuvant chemotherapy (20,21).
Other clinicopathological features prospectively shown to be independent, unfavorable prognostic factors in early NSCLC include older age, male sex and non-squamous-cell histology. Only performance status was likely to predict therapeutic effect (3,4,7). Likewise, certain histological subtypes, such as large-cell neuroendocrine carcinoma and pure bronchioloalveolar carcinoma which confer, respectively, worse or better outcome, could also guide treatment strategy (22,23). Interestingly, a high correlation between gene expression profile signatures and tumor histological phenotype has been shown for early NSCLC (24). It seems that even a thoroughly validated molecular signature does not outperform combined conventional clinical and pathologic variables in predicting survival of NSCLC patients (24). Therefore, incorporating the subtype and grade into conventional clinical models could provide predictive accuracy similar to that of well validated gene panels (24).
Molecular prognostic and predictive markers in early stage NSCLC
Gene expression profiling signatures
Gene expression profiles may facilitate treatment decisions in lung cancer, similar to their use to predict chemotherapy benefit in early stage breast cancer. Several groups have developed prognostic signatures based on mRNA, microRNA or proteomic profiles in order to better define patients with good prognosis who could potentially be spared adjuvant treatments, and those with poor prognosis who may benefit from successful adjuvant therapies. However, most prognostic signatures have some limitations that should be taken into account when analyzing their potential clinical utility. For instance, survival of NSCLC patients depends to a large extent on co-morbidity factors and the impact of this cannot be accounted for by prognostic tests based on tumor molecular profiling. At the same time, methodological or statistical data analyses have often been insufficient in the original studies proposing the signature for clinical use. Consider the recent example of the phase III Cancer and Leukemia Group B (CALGB) 30506 trial that was originally designed to validate the potential utility of a lung cancer metagene model in selecting patients with pT1 -T2N0 tumors for adjuvant chemotherapy. The study was recently amended as the original authors failed to replicate their own results. This highlights the need for very careful large-scale validation of prognostic signatures before they can be prospectively tested in clinical studies with adjuvant therapies for lung cancer (25).
Subramanian and Simon have recently published an elegant review of 16 published studies involving the analysis of gene expression data for developing prognostic signatures in NSCLC, in which they report serious methodological flaws in design and analysis, including inappropriate patient dataset selection, lack of independent validation, biased reporting of re-substitution statistics, incomplete protocol specification and use of statistical methods (26). Indeed, only a few gene signature studies have yielded data specifically referring to NSCLC stages IA, IB, or II that warrant further prospective validation (27-33). Among them it is worth mentioning the prognostic 15-gene signature for early NSCLC that was recently reported as the first deriving from prospectively collected tumor samples from patients enrolled in a phase III adjuvant trial. Gene expression profiling was conducted on mRNA from 133 frozen tumor samples from the National Cancer Institute of Canada Clinical Trials Group (NCIC CTG)-JBR.10 trial (28). The prognostic value of this gene signature was tested in four independent published microarray data sets and by quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR). Among these genes were nuclear proteins or transcription regulators such as mouse double minute 2 homolog (MDM2), zinc finger protein 236 (ZNF236), fos-related antigen 2 (FOSL2), hexamethylene bis-acetamide (HMBA)-inducible protein 1 (HEXIM1), myelin transcription factor 1-like (MYT1L) and inhibitor of kappa light polypeptide gene enhancer in B-cells (IKBKAP). The second subset of genes included protein melan-A (MLANA), ATPase subunit beta-1 (ATP1B1), L1 cell adhesion molecule (L1CAM), and stathmin-2 (STMN2), which encode for transmembrane- or membrane-associated proteins, potentially involved in signaling pathways and, finally, sodium/potassium-transporting ATPase subunit beta-1 (ATP1B1) and uridine monophosphate synthetase (UMPS) which are involved in purine and pyramidine metabolism, respectively, suggesting dependency of NSCLC on these pathways (28). This 15-gene expression profile was unique in that it could also predict response to systemic chemotherapy, whereas most other gene profiles have served only as prognostic markers following surgery. The signature was shown to interact significantly with the effect of cisplatin plus vinorelbine chemotherapy, with high-risk patients benefiting the most, although its potential predictive role requires independent validation. Also clinically relevant to the adjuvant strategy, this signature was able to assign, separately, stage IB and II patients to high- and low-risk subgroups with significantly different overall survival (28). When the predictive value of previously published prognostic signatures, applied to treated and untreated patients in the JBR.10 data set was evaluated, only the six-gene signature identified by Boutros was proved to be both significantly prognostic and predictive (27). The six-gene model comprised: syntaxin 1A (STX1A), hypoxia inducible factor 1A (HIF1A), chaperonin containing TCP1 subunit 3 (CCT3), MHC Class II DP beta 1 (HLA-DPB1), v-maf musculoaponeurotic fibrosarcoma onco-gene homolog K (MAFK), and ring finger protein 5 (RNF5) (27).
Finally, a 14-gene assay that uses RT-qPCR analysis of formalin-fixed, paraffin-embedded tissues was developed with a cohort of 361 patients with non-squamous NSCLC resected at the University of California, San Francisco, by Kratz, He, et al. from Michael Mann and David Jablons’ group (34). The investigators developed a 14-gene signature panel, consisting of 11 cancer-related genes: BCL2-associated athanogene (BAG1), breast cancer susceptibility gene 1 (BRCA1), cell division control protein 6 homolog (CDC6), cyclin-dependent kinase 2 associated protein 1 (CDK2AP1), receptor tyrosine-protein kinase erbB-3 (ERBB3), galactoside 3(4)-L-fucosyltransferas (FUT3), interleukin 11 (IL11), lymphocyte-specific protein tyrosine kinase (LCK), Rho family GTPase 3 (RND3), SH3 domain-binding glutamic acid-rich protein (SH3BGR), and wingless-type MMTV integration site family, member 3A (WNT3A) together with 3 reference genes, esterase D (ESD), TATA box binding protein (TBP) and Yes-associated protein 1 (YAP1). They validated the candidate gene signatures in 2 different populations: a community-based series of 433 resections for stage I non-squamous NSCLC from Northern California, and a cohort of 1,006 resections for stage IA-IIIB non-squamous NSCLC from the China Clinical Trials Consortium (34). The combination of gene signatures proved to be independently prognostic, irrespective of TNM stage grouping (34), in stage I, II and III patients. The prognostic value was significantly greater than certain clinical risk stratification criteria proposed by the US National Comprehensive Cancer Network for stage I resections (34). Furthermore, it was similarly effective in the Northern California and Chinese validation populations (34). However, there are some limitations to this study that should be pointed out. For instance, patients with squamous cell histology were excluded and there was poor overall quality of pathologic nodal staging, bearing in mind that 18% of resections for NSCLC in United States have no lymph nodes examined. As with all studies so far, this is another retrospective series, albeit the largest and most rigorously validated one performed to date.
Individual prognostic and predictive biomarkers in early-stage NSCLC
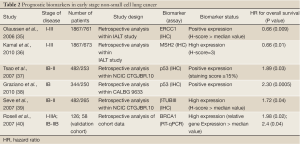
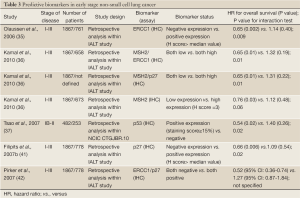
There are several candidate markers for sensitivity or resistance to chemotherapy identified in retrospective analyses of tumor biopsies from phase III clinical trials testing the value of adjuvant chemotherapy (Tables 2,3).
Full Table
Full Table
Excision repair cross complementation group 1 (ERCC1)
ERCC1 is a rate limiting enzyme in the nucleotide excision repair (NER) and interstrand cross-link repair pathways, which recognizes and repairs platinum induced adducts. Cancer cells overexpressing ERCC1 are more likely to have de novo resistance to cisplatin and a growing list of reports links cisplatin, carboplatin, and oxaliplatin resistance to ERCC1 mRNA levels in tumors. This relationship has been suggested for patients with gastric, bladder, ovarian, colorectal, and lung cancer. It was shown that ERCC1 levels evaluated by immunohistochemistry (IHC) are also predictive for the survival benefit afforded by adjuvant cisplatin-based chemotherapy in patients with totally resected stage I to IIIA NSCLC (35).
The International Adjuvant Lung Cancer Trial (IALT)-Bio translational research project aimed to study molecular biomarkers of tumors for their potential predictive values with regard to the effect of adjuvant chemotherapy on survival in IALT patients. Five groups of molecular biomarkers (19 markers in total) were studied by IHC: drug transporters, DNA repair, cell cycle regulators, signal transduction and apoptosis. Both ERCC1 and cyclin-dependent kinase inhibitor 1B (CDKN1B or p27) were found to have predictive value in patients with completely resected NSCLC undergoing adjuvant cisplatin-based chemotherapy (35). Interestingly, in patients randomly assigned to the observation arm, the subgroup with ERCC1-positive tumors had better survival compared with those with ERCC1-negative tumors (35). The paradoxical status of ERCC1, which was found to be a good prognostic marker in untreated resected NSCLC patients but a poor predictor of efficient adjuvant chemotherapy, was also confirmed in another study by Zheng et al, where the concomitant high expression of RRM1 and ERRC1 delineated a subgroup of chemonaïve patients with stage I disease with excellent survival outcomes (43). p27 is a tumor-suppressor protein that induces cell-cycle arrest in phase G1. Overexpression of p27 may confer de novo resistance to cisplatin by giving necessary time to repair cisplatin-induced DNA damage. In retrospective analysis of the IALT, patients with p27-negative tumors had longer survival after chemotherapy compared with surgery alone (41). Among six cell cycle regulators evaluated by IHC within the IALT-bio project, only p27 was identified to significantly correlate with treatment effect. Its predictive ability was independent from ERCC1 expression and, as anticipated, only patients with p27-negative tumors had survival benefit of cisplatin-based chemotherapy. Furthermore, when combining the IHC features of ERCC1 and p27, patients with tumors negative for both biomarkers seemed to benefit most from adjuvant chemotherapy (41).
In a pharmacogenomic trial with a biomarker-strategy design in advanced NSCLC by Cobo et al, ERCC1 mRNA expression was evaluated prospectively in an attempt to predict response to cisplatin-based or cisplatin-free chemotherapy regimens in stage IIIB or IV in NSCLC patients (44). Overall response rate was significantly higher in the genotypic arm, where chemotherapy regimen was tailored by ERCC1 mRNA expression (44). Patients in the control arm were not evaluated for the biomarker and received standard platinum-based combination (44). Within the customized arm, patients with low ERCC1 levels were treated with the same regimen as the control arm, whereas those with high levels received a non platinum regimen (44). Most importantly, however, clinical relevance remained limited, given that there was no difference between the two arms in either progression-free survival or overall survival. This paradox of favorable long term outcome despite cisplatin chemoresistance probably indicates that, by preventing mutagenesis, DNA repair may not only prevent cancer but may retard molecular events related to progression in established tumors. Thus, high expression of ERCC1 may indicate a favorable outcome in these untreated patients by identifying tumors that have progressed relatively little at the molecular level. Intact DNA repair mechanisms prevent accumulation of genetic aberrations that confer a high malignant potential (45). A recent meta-analysis failed to support two common ERCC1 gene polymorphisms ERCC1 C118T/C8092A and ERCC2 Lys751Gln/Asp312Asn as useful prognostic factors for assessing treatment response to platinum-based chemotherapies in NSCLC patients (46).
Additional biomarkers related to the repair of cisplatin-induced DNA damage have been included in the IALT bio project in order to enhance the predictive power of ERCC1. MutS homolog 2 (MSH2) is a major active component of the mismatch repair machinery; IHC expression of MSH2 displayed a very similar pattern of significance to ERCC1. Specifically, patients with low MSH2 levels had markedly better survival with adjuvant chemotherapy. Those with high levels seemed to have no benefit and in fact had a median survival 9 months shorter than those who did not receive chemotherapy, although this was not statistically significant. Similar to the prognostic role of other DNA-excision-repair proteins, high MSH2 levels predicted significantly longer survival in patients in the observation arm. When MSH2 and ERCC1 expression patterns were combined to form four phenotypes, the benefit from chemotherapy was significantly greater for patients with double-negative tumors. This was also noted when MSH2 expression was combined with that of p27, suggesting that MSH2 immunostaining was a superior predictive biomarker when considered jointly with either of the two other variables (36).
Ribonucleotide reductase subunit M1 (RRM1)
RRM1 is a regulatory component of ribonucleoside-diphosphate reductase, a key enzyme in DNA synthesis that catalyzes the formation of deoxyribonucleotides, by reducing ribonucleotides. The reaction requires generation of a radical allowing the 2'-hydroxyl of ribose to be reduced, which is carried out by the RRM1 enzyme (47). The antimetabolite gemcitabine interferes with the function of RRM1 by reducing the pool of deoxyribonucleotide-5'-diphosphate available for DNA synthesis (45). Although relevant data for the adjuvant setting are lacking, correlative studies within randomized clinical trials in advanced NSCLC have shown that RRM1 overexpression, either at the mRNA or protein level, predicts poor response to gemcitabine-based chemotherapy (48-50). It is somewhat surprising that RRM1 protein expression has recently predicted outcome in patients treated with cisplatin and vinorelbine in a biomarker study. In this treatment arm, patients without RRM1 protein expression showed improved disease control rates, progression-free survival and overall survival, while RRM1 had no predictive impact in patients treated with cisplatin, paclitaxel and gemcitabine (51).
In contrast to predicting chemoresistance, RRM1 is a biologically and clinically important determinant of malignant behavior in NSCLC whose overexpression seems to confer favorable outcome. RRM1 suppresses cell migration and metastasis, which is at least partially mediated through induction of the phosphatase and tensin homolog gene (PTEN). RRM1 is in a region of frequent loss of heterozygosity (LOH), and LOH at this locus was found to be an independent determinant of poor survival in a large cohort of patients with stage I and II NSCLC (52). High RRM1 transcriptional expression, defined as mRNA levels above the median value, was favorably prognostic of survival in two independent cohorts of patients with resected NSCLC, most of who were diagnosed at early stage and treated with surgery only. In this study, RRM1 overexpression was a stage-independent predictor of survival, albeit highly correlated with PTEN expression (53). Longer overall survival was recently found in another group of NSCLC patients with high RRM1 mRNA expression who had undergone curative lung resection (54). The prognostic role of RRM1 was also confirmed by Zheng et al. who measured RNA expression of RRM1 and ERCC1 using RT-qPCR in fresh frozen and formalin fixed paraffin-embedded tumor samples (43). This study showed that RRM1 expression correlated with ERCC1 expression and that patients whose tumors had high expression of RRM1 had superior survival compared with the low expression group (43). In contrast to the previous study, there was no correlation with PTEN expression at the protein level. Interestingly, the concomitant high expression of RRM1 and ERCC1 delineated a subgroup of patients with excellent survival outcomes, accounting for 30% of the cohort (43).
Breast cancer susceptibility gene 1 (BRCA1)
BRCA1 is a multifunctional nuclear phosphoprotein which is ubiquitously expressed in all tissues and serves in part as a tumor suppressor, a “caretaker” and a “gatekeeper” in preserving genomic stability. BRCA1 has recently emerged as one of the most appealing biomarkers for personalizing chemotherapy in NSCLC. It has been implicated in normal cellular functions including cell cycle regulation, replication, mitotic spindle assembly, transcription regulation and higher chromatin hierarchical control (55). Also, BRCA1 has a crucial role in DNA repair as a component of the transcription-coupled NER and the homologous recombinant repair pathways. BRCA1 functions as a sensitizer to apoptosis induced by antimicrotubulin agents, such as taxanes and vinca alkaloids and also abrogates apoptosis induced by a range of DNA-damaging agents, including cisplatin and etoposide. Upstream activity of the receptor-associated protein 80 (RAP-80) is required for localization of BRCA1 to sites of DNA double-strand breaks (55).
In a recently reported feasibility study, adjuvant chemotherapy was customized based on BRCA1 mRNA levels in 84 patients with completely resected NSCLC. Patients with higher BRCA1 transcriptional levels were treated with single agent docetaxel, whereas those with intermediate and low BRCA1 expression received cisplatin-based doublets. Interim analyses showed that single-agent docetaxel was not inferior to cisplatin/docetaxel in terms of survival in patients with high BRCA1 levels (56). Therefore, high BRCA1 predicts resistance to cisplatin and possibly sensitivity to docetaxel. Expression levels of BRCA1, divided in quartiles, were assessed in a cohort of 55 patients with stage II to IIIA NSCLC who received neoadjuvant chemotherapy with cisplatin/gemcitabine followed by complete resection. Those with the lowest levels of BRCA1 mRNA expression had significantly greater benefit from chemotherapy in terms of clinical and pathological downsizing as well as overall survival (57).
The potential prognostic role of BRCA1 was investigated in two independent cohorts of chemonaïve patients with early-stage NSCLC analyzed by RT-qPCR. In the study by Rosell et al., expression level of nine genes involved in DNA repair, including BRCA1, were correlated with overall survival in 126 NSCLC patients who had undergone complete resection and did not receive adjuvant chemotherapy (40). In a univariate analysis, three genes appeared to influence relapse: myeloid zinc finger 1 (MZF1), thioredoxin-1 (TRX1), and BRCA1. However, only BRCA1 and stage III disease remained significant predictors of survival in the multivariate analysis (40). For the 40 patients with a high level of BRCA1 expression, median survival was 29 months while median survival was not reached for the 83 patients with low BRCA1 expression (40). The striking lack of prognostic significance of other biomarkers included in this study may be partially due to the strong intergene coexpression, such as that between BRCA1 and ERCC1, observed. The independent adverse prognostic effect of high BRCA1 expression was confirmed in another cohort of patients with early stage NSCLC also evaluated for ERCC1 and RRM1 mRNA levels. In this study, xeroderma pigmentosum complementation group G (XPG), a key gene for the NER system, was identified as an independent favorable predictor of survival outcome, as well as a potential modulator of recurrence risk among patients with BRCA1 overexpression (58).
Thymidylate synthase (TS)
Thymidylate synthase (TS) and methylenetetrahydrofolate reductase (MTHFR) play important roles in folate metabolism. TS is an enzyme involved in purine synthesis and, as an anti-cancer chemothera¬py target, can be inhibited by TS inhibitors such as fluorinated pyrimidine fluorouracil or certain folate analogues, most notably pemetrexed. Consistent findings across phase III trials in advanced NSCLC have established the favorable predictive effect of non-squamous cell histology on treatment with pemetrexed (59). Differentially high TS expression in squamous cell NSCLC represents the main molecular basis underlying this treatment by histology interaction. Data from a current study indicate higher TS expression levels in squamous cell and in high-grade carcinomas (60). No clinical data exist to confirm the predictive role of either histology or TS expression in the adjuvant setting. However an independent prognostic effect for TS has been revealed in chemonaïve patients with resected early stage NSCLC. TS protein expression was correlated significantly with higher proliferative activity of NSCLC cells and, consequently, with poor prognosis in patients with NSCLC who had higher TS level (61). Two other different studies of chemonaïve patients with resected early stage NSCLC revealed an independent prognostic effect for TS, but with conflicting qualitative results. High TS expression at the mRNA, but not IHC level, was significantly associated with adverse disease-free survival in the study from Shintani et al. High TS expression as determined by automated in situ protein quantification, but not by RT-qPCR, predicted improved overall survival in the latter study, in which also TS protein levels did not correlate with those of ERCC1 and RRM1 (62,63). No correlations between intratumoral TS levels and any known clinicopathological variables were reported, with the exception of a recently published article in which TS gene expression was associated with disease stage, lymph node metastasis, tumor differentiation, prognosis, and tumor cell proliferation in patients with lung adenocarcinoma (64).
Kirsten-rous avian sarcoma (KRAS) and p53
The Kirsten-rous avian sarcoma (KRAS) protein is a member of the RAS family of proteins that encode small GTPases involved in cellular signal transduction. Activation of Ras signalling causes cell growth, differentiation, and survival, by transmitting signal downstream from growth factor receptors, including epidermal growth factor receptor (EGFR). From the three RAS genes, KRAS contains > 90% of the mutations detectable in almost 10-25% of NSCLC and mostly affecting codon 12 and 13 (65). Recent data suggested that KRAS mutations may affect outcome of NSCLC patients receiving chemotherapy. In the adjuvant setting, data from the JBR10 trial suggested no benefit from adjuvant chemotherapy in KRAS mutated patients (3,37). Similarly, a retrospective analysis of patients with stage IB disease enrolled in the phase III CALGB-9633 study showed that, among those with tumors larger than 4 cm, KRAS mutations may predict less overall survival benefit from the combination of carboplatin plus paclitaxel (66). In the LACE-BIO pooled analysis, the prognostic and predictive role of KRAS mutations was investigated in 1751 patients treated with adjuvant chemotherapy. Among evaluable patients, KRAS mutations had no effect on survival (67). It should be stressed that the formal test for interaction between the biomarker and treatment effect was not statistically significant in any of the above studies. KRAS mutation status is associated with cigarette smoking and adenocarcinoma histology. The role of KRAS mutations as a prognostic factor in NSCLC remains controversial. Although some studies suggested a potential negative prognostic effect, other studies did not confirm any negative impact on survival for individuals harboring KRAS mutation (68-71).
The TP53 gene, located on the short arm of human chromosome 17, encodes for a nuclear phosphoprotein involved in the regulation of cell proliferation. The tumor suppressor protein, p53, has a wide range of functions, most of which are mediated via regulation of gene transcription. Commonly described as ‘the genome guardian’, p53 is involved in important cellular processes, such as stress response, cell-cycle control, DNA repair and apoptosis. The mutant gene product, which tends to accumulate to high levels in cancer cells, is believed to exert a dominant negative effect over coexpressed normal TP53. In resected lung cancers, point mutations of the TP53 gene have been found in all histologic types, including approximately 45% of resected NSCLC and, even more frequently, in small-cell lung cancer. Similar to observations with KRAS mutations, p53 mutations have been retrospectively correlated with clinical features such as younger age and squamous histology, but not sex, tumor stage, nodal status, neuroendocrine differentiation, or prior chemotherapy. Many retrospective studies have examined the prognostic role of p53 gene mutations in NSCLC. However, most of these studies have been limited by small size, heterogeneous patient samples, potential selection biases, and/or insensitive p53 mutation detection techniques, leading to inconsistent results. p53 mutations have been associated with decreased survival, no statistically significant change in survival, or improved survival in NSCLC. Previous meta-analyses have indicated that TP53 mutations and p53 expression are weak predictors of outcome in NSCLC (72,73). In contrast, the first published prospective trial examining the prognostic role of p53 mutations in NSCLC demonstrated that neither p53 expression nor TP53 mutations were shown to have predictive value. However, this should be interpreted taking into consideration the different study design and the use of an old-generation regimen (74). However, in the retrospective companion analysis of the phase III NCIC CTG-JBR.10 adjuvant trial p53 IHC overexpression was found to be an independent unfavorable prognostic factor among patients in the observation arm. In addition, only patients with p53-positive tumors derived benefit from the cisplatin plus vinorelbine combination. In contrast to p53 expression, TP53 mutation status was neither prognostic for survival, nor predictive for efficacy of adjuvant chemotherapy (37). This suggests that the biological effects of TP53 mutations and p53 protein overexpression are not equal, highlighting their complex role in tumor aggressiveness and chemosensitivity. Finally, a recent biomarker study as part of the phase III CALGB- 9633 adjuvant trial identified p53 and mucin overexpression as independent adverse prognostic factors for stage IB patients (38).
β-TUBULIN and epidermal growth factor receptor
β-tubulin is an essenti al element of microtubules, which, in turn, serve as cellular structural components involved in vital processes, including mitosis. Among described mechanisms of resistance to anti-tubulin agents, class III β-tubulin (βTUBIII) overexpression is of particular interest. To assess whether βTUBIII might be a useful marker in early NSCLC patients undergoing adjuvant chemotherapy with a vinorelbine-based regimen, levels of βTUBIII were measured in tumor samples from patients treated in the NCIC CTG JBR.10 study. No significant interaction between the biomarker and the effect of cisplatin plus vinorelbine combination was found. Subgroup analysis suggested that high, rather than low, βTUBIII levels were predictive for chemotherapy benefit. However, high βTUBIII expression was shown to be an independent adverse predictor of recurrence- free survival (39). Its prognostic value was retrospectively confirmed in patients enrolled in the IALT study (75). The adverse prognostic significance of high βTUBIII expression is consistent with prior published reports in the setting of advanced NSCLC. Rosell et al. correlated high βTUBIII mRNA levels with inferior outcome in advanced NSCLC patients treated with anti-tubulin agents (76). It has also been shown that high level of expression of βTUBIII in tumor cells, assessed by a semiquantitative IHC assay, was associated with a lower response rate and poor prognosis in advanced NSCLC patients receiving vinorelbine-based chemotherapy (77). In a recent study, high tumor expression of βTUBIII, assessed by IHC in 47 NSCLC patients receiving a paclitaxel-based regimen, was predictive of lower response to therapy and inferior survival (78).
Epidermal growth factor receptor (EGFR) status, defined by mutation analysis or amplification by fluorescent in situ hybidization (FISH), was recently explored in correlation with the results of the phase III NCIC CTG-JBR.10 adjuvant trial. Neither sensitizing mutations nor high gene copy were significantly prognostic in the observation arm. Similarly, although there was a trend toward greater benefit from the cisplatin plus vinorelbine combination, interaction between the biomarkers and treatment effect was not significant (79). After adjusting for covariates, a recent large, prospective, cohort study of patients with stage I-III adenocarcinoma, 20% of whom had received perioperative chemotherapy, failed to show any significant association between overall survival and mutation status of either EGFR or KRAS (80). In conclusion, the potential prognostic and predictive effect of EGFR amplification and the two most prominent mutations, del 19 and L858R, with regard to chemotherapy effect in the adjuvant setting remains undefined.
Conclusions
Finally, are we ready to adopt the use of biomarkers into early stage NSCLC staging, prognosis and treatment selection? Survival amongst cancer patients has improved in recent decades with the availability and application of various treatment modalities. Tumor classification, stage and, sometimes, grade are used to assess prognosis. Although adjuvant chemotherapy has been well established for patients with early stage NSCLC, stage alone is not an ideal biomarker to predict the utility of chemotherapy as the vast majority of patients derive no benefit from treatment. The discovery of molecular biomarkers with the potential to select high-risk patients and predict drug efficacy is essential, especially in controversial fields such as treatment of elderly patients and stage I disease. Biomarker expression often supplants or complements tumor classification, stage or grade. In recent years, a widespread search for new, tumor biology-driven therapeutics has begun, especially in advanced NSCLC. However in the adjuvant setting, it seems that discovery of so-called promising markers translates rather slowly into clinical applicability and few markers have so far been integrated into clinical practice. There are many practical issues, such as the pharmaceutical companies concerns regarding fractionation of markets and medico-legal fears surrounding generation and possession of information. Furthermore, it can take time for physicians and patients to accept and adopt customizing adjuvant chemotherapy. However, as therapies become increasingly target specific, biomarkers will inevitably develop in tandem to play greater roles in staging, grading, and selection of adjuvant therapy; the practical hurdles are many and complex. As already mentioned, the RT-qPCR strategy involving a relatively small number of gene biomarkers and the use of paraffin-embedded specimens seems to outperform wide-genome profiling, although cutoff point definition for continuous variables, such as transcript levels, is particularly challenging because of the great inter-individualization variation of gene expression. In addition, the limited size of most studies and variable techniques used for marker determination plays a role. Often, initially promising results are not reproducible. Another important point is the possible discordance of biomarker status between different types of assays and the corresponding differences in association with clinical outcomes. As already discussed, mRNA expression of a biomarker gene does not necessarily correlate with protein levels as determined by IHC. Apart from multiple technical issues that potentially affect the results of each method, biomarker expression at the protein level depends on additional translational factors, such as microRNA, posttranslational modifications and degradation.
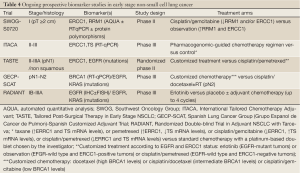
Although the cisplatin plus vinorelbine doublet is currently the standard option for adjuvant chemotherapy, use of appropriate surrogate biomarkers would facilitate randomized clinical trials to establish alternative or superior regimens with smaller sample sizes and shorter follow-up time. The prognostic and/or predictive role of many of the aforementioned biomarkers has been strongly supported by retrospective translational studies. Ideally, biomarkers should be validated analogously in prospective, well-controlled clinical studies of diverse patients across multiple institutions, with well-established standards for all steps of the process. We are anxiously awaiting the results of four prospective multicentre clinical trials of customized adjuvant strategy currently underway (Table 4). ERCC1, RRM1 and BRCA1 are considered to be among the most promising biomarkers with stage-independent, combined prognostic and predictive value, the clinical utility of which is being validated in the ongoing large-scale, randomized phase II and III trials. Until the, highly anticipated, results are in, neither these nor other candidate biomarkers should be used in daily clinical practice as decision-making criteria.
Full Table
These steps towards personalized medicine will hopefully represent a shift in the management of early staged NSCLC. Indeed, NSCLC should no longer be viewed as one common generic disease, but rather as a collection of rarer tumors with differing biological behaviors and sensitivities to various systemic treatments.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Scagliotti GV, Fossati R, Torri V, et al. Randomized study of adjuvant chemotherapy for completely resected stage I, II, or IIIA non-small-cell Lung cancer. J Natl Cancer Inst 2003;95:1453-61. [PubMed]
- Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol 2010;28:35-42. [PubMed]
- Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. [PubMed]
- Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association (ANITA)): a randomised controlled trial. Lancet Oncol 2006;7:719-27. [PubMed]
- Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 2008;26:5043-51. [PubMed]
- Waller D, Peake MD, Stephens RJ, et al. Chemotherapy for patients with non-small cell lung cancer: the surgical setting of the Big Lung Trial. Eur J Cardiothorac Surg 2004;26:173-82. [PubMed]
- Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. [PubMed]
- Crinò L, van Meerbeeck J. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21:v103-15. [PubMed]
- Pisters KM, Evans WK, Azzoli CG, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I-IIIA resectable non small-cell lung cancer guideline. J Clin Oncol 2007;25:5506-18. [PubMed]
- Robinson LA, Ruckdeschel JC, Wagner H Jr, et al. Treatment of non-small cell lung cancer-stage IIIA: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 2007;132:243S-265S.
- Scott WJ, Howington J, Feigenberg S, et al. Treatment of non-small cell lung cancer stage I and stage II: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 2007;132:234S-242S.
- Pocock SJ, Assmann SE, Enos LE, et al. Subgroup analysis, covariate adjustment and baseline comparisons in clinical trial reporting: current practice and problems. Stat Med 2002;21:2917-30. [PubMed]
- Hamada C, Tanaka F, Ohta M, et al. Meta-analysis of postoperative adjuvant chemotherapy with tegafur-uracil in non-small-cell lung cancer. J Clin Oncol 2005;23:4999-5006. [PubMed]
- Rami-Porta R, Ball D, Crowley J, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol 2007;2:593-602. [PubMed]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. [PubMed]
- Strauss GM, Wang XF, Maddaus M, et al. Adjuvant chemotherapy (AC) in stage IB non-small cell lung cancer (NSCLC): Long-term follow-up of Cancer and Leukemia Group B (CALGB) 9633. ASCO Meeting Abstracts 29:7015. OpenURL.
- Varlotto JM, Recht A, Nikolov M, et al. Extent of lymphadenectomy and outcome for patients with stage I nonsmall cell lung cancer. Cancer 2009;115:851-8. [PubMed]
- Jonnalagadda S, Arcinega J, Smith C, et al. Validation of the lymph node ratio as a prognostic factor in patients with N1 nonsmall cell lung cancer. Cancer 2011;117:4724-31. [PubMed]
- Jonnalagadda S, Smith C, Mhango G, et al. The number of lymph node metastases as a prognostic factor in patients with N1 non-small cell lung cancer. Chest 2011;140:433-40. [PubMed]
- Ruffini E, Asioli S, Filosso PL, et al. Significance of the presence of microscopic vascular invasion after complete resection of Stage I-II pT1-T2N0 non-small cell lung cancer and its relation with T-Size categories: did the 2009 7th edition of the TNM staging system miss something? J Thorac Oncol 2011;6:319-26.
- Maeda R, Yoshida J, Ishii G, et al. Poor prognostic factors in patients with stage IB non-small cell lung cancer according to the seventh edition TNM classification. Chest 2011;139:855-61.
- Travis WD, Garg K, Franklin WA, et al. Bronchioloalveolar carcinoma and lung adenocarcinoma: the clinical importance and research relevance of the 2004 World Health Organization pathologic criteria. J Thorac Oncol 2006;1:S13-19. [PubMed]
- Iyoda A, Hiroshima K, Nakatani Y, et al. Pulmonary large cell neuroendocrine carcinoma: its place in the spectrum of pulmonary carcinoma. Ann Thorac Surg 2007;84:702-7. [PubMed]
- Sun Z, Yang P. Gene expression profiling on lung cancer outcome prediction: present clinical value and future premise. Cancer Epidemiol Biomarkers Prev 2006;15:2063-8. [PubMed]
- Potti A, Mukherjee S, Petersen R, et al. Retraction: A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med 2006;355:570-80. N Engl J Med 2011;364:1176. [PubMed]
- Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst 2010;102:464-74. [PubMed]
- Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A 2009;106:2824-8. [PubMed]
- Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol 2010;28:4417-24. [PubMed]
- Kadara H, Behrens C, Yuan P, et al. A five-gene and corresponding protein signature for stage-I lung adenocarcinoma prognosis. Clin Cancer Res 2011;17:1490-501. [PubMed]
- Broët P, Camilleri-Broet S, Zhang S, et al. Prediction of clinical outcome in multiple lung cancer cohorts by integrative genomics: implications for chemotherapy selection. Cancer Res 2009;69:1055-62. [PubMed]
- Roepman P, Jassem J, Smit EF, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res 2009;15:284-90. [PubMed]
- Bianchi F, Nuciforo P, Vecchi M, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest 2007;117:3436-44. [PubMed]
- Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med 2006;3:e467. [PubMed]
- Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet 2012;379:823-32. [PubMed]
- Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [PubMed]
- Kamal NS, Soria JC, Mendiboure J, et al. MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clin Cancer Res 2010;16:1206-15. [PubMed]
- Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol 2007;25:5240-7. [PubMed]
- Graziano SL, Gu L, Wang X, et al. Prognostic significance of mucin and p53 expression in stage IB non-small cell lung cancer: a laboratory companion study to CALGB 9633. J Thorac Oncol 2010;5:810-7. [PubMed]
- Sève P, Lai R, Ding K, et al. Class III beta-tubulin expression and benefit from adjuvant cisplatin/vinorelbine chemotherapy in operable non-small cell lung cancer: analysis of NCIC JBR.10. Clin Cancer Res 2007;13:994-9. [PubMed]
- Rosell R, Skrzypski M, Jassem E, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One 2007;2:e1129. [PubMed]
- Filipits M, Pirker R, Dunant A, et al. Cell cycle regulators and outcome of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer: the International Adjuvant Lung Cancer Trial Biologic Program. J Clin Oncol 2007;25:2735-40. [PubMed]
- Pirker R, Filipits M, Dunant A, et al. Ialt-Bio: A challenging research to improve adjuvant chemotherapy of completely resected NSCLC: D3-03. J Thorac Oncol 2007;2:S397-S398.
- Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med 2007;356:800-8. [PubMed]
- Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol 2007;25:2747-54. [PubMed]
- Gazdar AF. DNA repair and survival in lung cancer--the two faces of Janus. N Engl J Med 2007;356:771-3. [PubMed]
- Yin M, Yan J, Voutsina A, et al. No evidence of an association of ERCC1 and ERCC2 polymorphisms with clinical outcomes of platinum-based chemotherapies in non-small cell lung cancer: a meta-analysis. Lung Cancer 2011;72:370-377. [PubMed]
- Poole AM, Logan DT, Sjoberg BM. The evolution of the ribonucleotide reductases: much ado about oxygen. J Mol Evol 2002;55:180-96. [PubMed]
- Reynolds C, Obasaju C, Schell MJ, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol 2009;27:5808-15. [PubMed]
- Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res 2004;10:1318-25. [PubMed]
- Boukovinas I, Papadaki C, Mendez P, et al. Tumor BRCA1, RRM1 and RRM2 mRNA expression levels and clinical response to first-line gemcitabine plus docetaxel in non-small-cell lung cancer patients. PLoS One 2008;3:e3695. [PubMed]
- Vilmar AC, Santoni-Rugiu E, Sorensen JB. Predictive impact of RRM1 protein expression on vinorelbine efficacy in NSCLC patients randomly assigned in a chemotherapy phase III trial. Ann Oncol 2013;24:309-14. [PubMed]
- Bepler G, Gautam A, McIntyre LM, et al. Prognostic significance of molecular genetic aberrations on chromosome segment 11p15.5 in non-small-cell lung cancer. J Clin Oncol 2002;20:1353-60. [PubMed]
- Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol 2004;22:1878-85. [PubMed]
- Pesta M, Kulda V, Fiala O, et al. Prognostic significance of ERCC1, RRM1 and BRCA1 in surgically-treated patients with non-small cell lung cancer. Anticancer Res 2012;32:5003-10. [PubMed]
- Reguart N, Cardona AF, Carrasco E, et al. BRCA1: a new genomic marker for non-small-cell lung cancer. Clin Lung Cancer 2008;9:331-9. [PubMed]
- Cobo M, Massuti B, Moran T, et al. Spanish customized adjuvant trial (SCAT) based on BRCA1 mRNA levels. ASCO Meeting Abstracts 2008;26:7533.
- Taron M, Rosell R, Felip E, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet 2004;13:2443-9. [PubMed]
- Bartolucci R, Wei J, Sanchez JJ, et al. XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung Cancer 2009;10:47-52. [PubMed]
- Scagliotti G, Hanna N, Fossella F, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two Phase III studies. Oncologist 2009;14:253-63. [PubMed]
- Ceppi P, Volante M, Saviozzi S, et al. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase. Cancer 2006;107:1589-96. [PubMed]
- Nakagawa T, Otake Y, Yanagihara K, et al. Expression of thymidylate synthase is correlated with proliferative activity in non-small cell lung cancer (NSCLC). Lung Cancer 2004;43:145-9. [PubMed]
- Shintani Y, Ohta M, Hirabayashi H, et al. New prognostic indicator for non-small-cell lung cancer, quantitation of thymidylate synthase by real-time reverse transcription polymerase chain reaction. Int J Cancer 2003;104:790-5. [PubMed]
- Zheng Z, Li X, Schell MJ, et al. Thymidylate synthase in situ protein expression and survival in stage I nonsmall-cell lung cancer. Cancer 2008;112:2765-73. [PubMed]
- Nakagawa T, Tanaka F, Otake Y, et al. Prognostic value of thymidylate synthase expression in patients with p-stage I adenocarcinoma of the lung. Lung Cancer 2002;35:165-70. [PubMed]
- Karachaliou N, Mayo C, Costa C, et al. KRAS Mutations in Lung Cancer. Clin Lung Cancer 2013;14:205-14. [PubMed]
- Capelletti M, Wang XF, Gu L, et al. Impact of KRAS mutations on adjuvant carboplatin/paclitaxel in surgically resected stage IB NSCLC: CALGB 9633. J Clin Oncol 2010;28:7008.
- M.S. Tsao PH, Bourredjem A, et al. LACE-bio pooled analysis of the prognostic and predictive value of KRAS mutation in completely resected non-small cell lung cancer (NSCLC).Ann Oncol 2010;21 supple 8, abstract 4218.
- Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990;323:561-5. [PubMed]
- Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92:131-9. [PubMed]
- Mitsudomi T, Steinberg SM, Oie HK, et al. ras gene mutations in non-small cell lung cancers are associated with shortened survival irrespective of treatment intent. Cancer Res 1991;51:4999-5002. [PubMed]
- Grossi F, Loprevite M, Chiaramondia M, et al. Prognostic significance of K-ras, p53, bcl-2, PCNA, CD34 in radically resected non-small cell lung cancers. Eur J Cancer 2003;39:1242-50. [PubMed]
- Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J 2001;18:705-19. [PubMed]
- Mitsudomi T, Hamajima N, Ogawa M, et al. Prognostic significance of p53 alterations in patients with non-small cell lung cancer: a meta-analysis. Clin Cancer Res 2000;6:4055-63. [PubMed]
- Schiller JH, Adak S, Feins RH, et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol 2001;19:448-57. [PubMed]
- Rosell R, Scagliotti G, Danenberg KD, et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene 2003;22:3548-53. [PubMed]
- Sève P, Isaac S, Trédan O, et al. Expression of class III {beta}-tubulin is predictive of patient outcome in patients with non-small cell lung cancer receiving vinorelbine-based chemotherapy. Clin Cancer Res 2005;11:5481-6. [PubMed]
- Sève P, Mackey J, Isaac S, et al. Class III beta-tubulin expression in tumor cells predicts response and outcome in patients with non-small cell lung cancer receiving paclitaxel. Mol Cancer Ther 2005;4:2001-7. [PubMed]
- Tsao MS, Sakurada A, Ding K, et al. Prognostic and predictive value of epidermal growth factor receptor tyrosine kinase domain mutation status and gene copy number for adjuvant chemotherapy in non-small cell lung cancer. J Thorac Oncol 2011;6:139-47. [PubMed]
- D’Angelo SP, Janjigian YY, Kris MG, et al. Impact of EGFR and KRAS mutations on survival in 1,000 patients with resected lung adenocarcinoma. J Clin Oncol 2010;28:7011.


